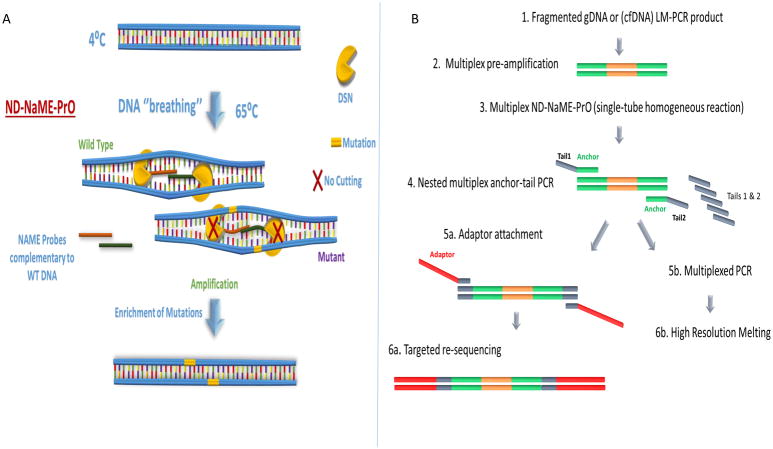
Figure 1.
Concept and workflow for No-Denaturation Nuclease-assisted Minor-allele Enrichment using PRobe-Overlap, ND-NaME-PrO. A. ND-NaME-PrO the spontaneous partial denaturation of dsDNA (DNA ‘breathing’) at elevated temperatures remaining below DNA melting temperatures, allowing overlapping probes to bind to complementary target DNA strands. Double strand specific nuclease DSN is then digesting fully matched templates while mismatched sequences remain substantially undigested, thereby resulting to mutation enrichment upon subsequent amplification. It should be noted that dsDNA not targeted by probes also becomes digested, but at a much lower rate than targeted DNA (vide infra). B. Sample preparation workflow for multiplexed ND-NaME-PrO followed by targeted re-sequencing. The workflow includes an initial multiplex amplification from genomic DNA or cfDNA followed by nested multiplex anchor-tail PCR for a target panel. The anchor-tail-PCR product is then enriched for mutations at multiple positions via multiplexed ND-NaME-PrO, followed by multiplexed HRM-scanning and MiSeq sequencing of HRM-positive samples.