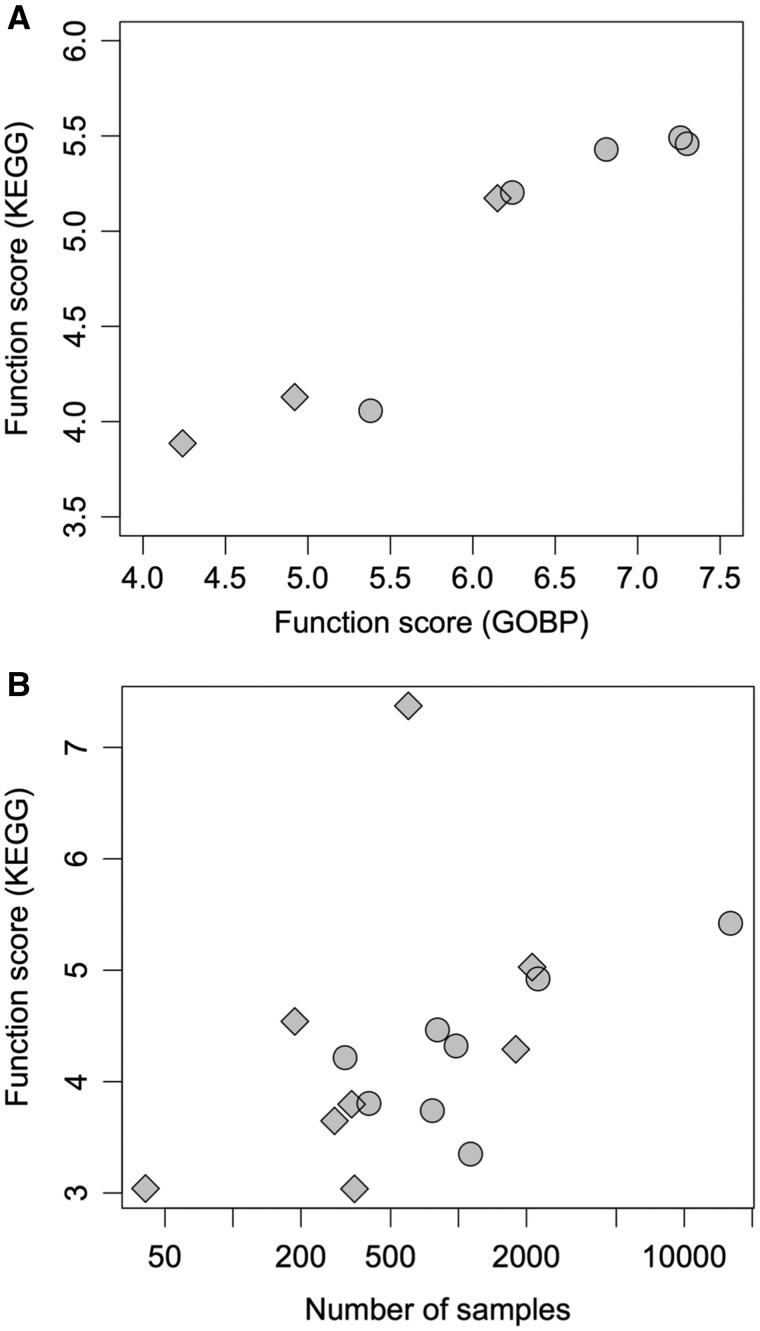
Fig. 1.
Quality assessment based on the consistency of known gene functions. As a measure of the quality of gene coexpression data, the power to discriminate gene pairs sharing a common functional annotation from other gene pairs was used. (A) Previous and current Arabidopsis coexpression data were assessed by this discrimination analysis. Irrespective of the gene annotation source (GO Biological Process or KEGG pathway), the quality trend was consistent. (B) The current 16 coexpression platforms were assessed with the discrimination analysis using the KEGG pathway annotation. Circles and diamonds indicate microarray-based and RNAseq-based coexpression datasets, respectively.