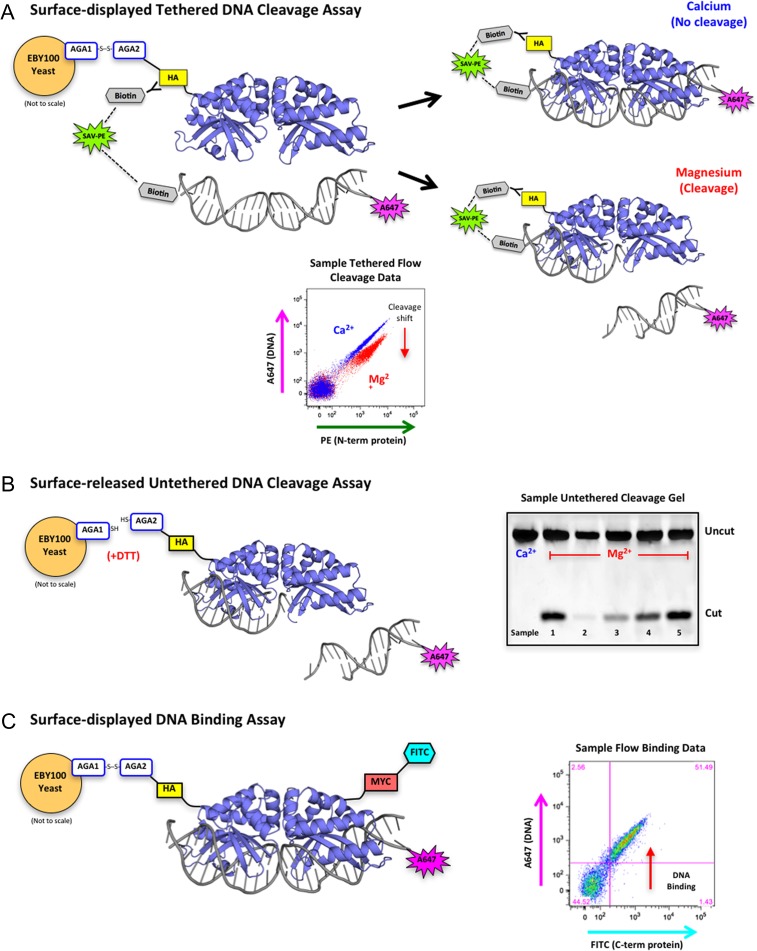
Fig. 1.
Overview of yeast surface display and flow cytometric assays. (A) Schematic of the tethered flow cytometric cleavage assay. A meganuclease is expressed on the surface of yeast with an N-terminal HA epitope tag. The HA tag is stained with a biotinylated anti-HA antibody and used to create a physical tether between the protein and a fluorescently labeled DNA target substrate via a fluorescent streptavidin-phycoerythrin (SAV-PE) bridge. In the presence of calcium, the DNA is able to bind but not cleave (blue diagonal population on the sample flow data plot). In the presence of magnesium, the DNA may be cleaved (red population in sample flow plot), which releases the fluorescent tag and produces a drop in A647 signal (cleavage shift designated by the red arrow). The ratio of A647 signal in the calcium vs. magnesium samples is used to quantitate cleavage activity against a given DNA target substrate. (B) Schematic of the surface-released untethered cleavage assay. The meganuclease is released from the surface of the yeast with dithiothreitol (DTT) and the A647-labeled DNA substrate is free-floating in solution with no tethering. This assay requires the enzyme to both bind and cleave the DNA substrate before cleavage can occur. The cleaved products are separated on an acrylamide gel and visualized by the fluorescence of the A647 tag on the DNA substrate. The sample untethered cleavage gel displays cleaved products from five unique samples with varying levels of cleavage. (C) Schematic of the flow cytometric DNA binding assay. The meganuclease is expressed on the surface of yeast, stained with an anti-Myc-FITC antibody to detect full-length protein expression (the Myc epitope tag is located at the C-terminal end of the protein), and incubated with varying concentrations of untethered DNA target substrate. Cells with both FITC and A647 signal (see sample flow binding plot) are expressed on the yeast surface and have bound the DNA substrate