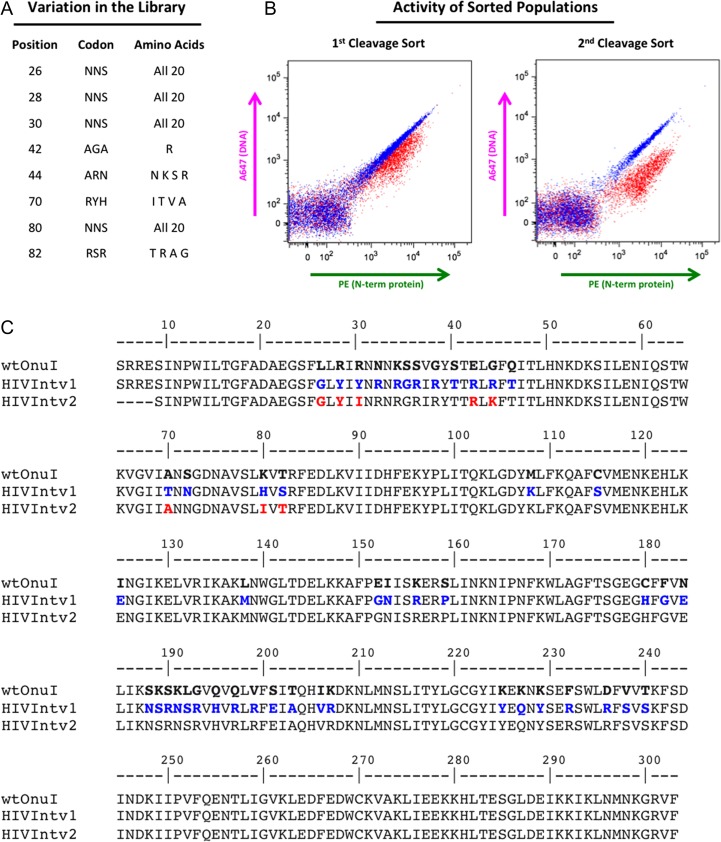
Fig. 6.
eOnu_HIVInt meganuclease engineering. (A) Eight amino acid positions were considered for variation in our eOnu_HIVInt engineering library. The codon used and variation introduced at each position is listed. (B) Cleavage of the desired HIVInt target sequence by sorted yeast populations is maintained and improved from the first to the second library sorting step, as illustrated by the magnitude of A647 shifts in the tethered flow cytometric cleavage assay. (C) Amino acid sequences are shown for wild type I-OnuI (the parent engineering scaffold), the original eOnu_HIVInt_v1 meganuclease (HIVIntv1), and our further engineered eOnu_HIVInt_v2 meganuclease (HIVIntv2). Numbering of residues matches that of the original I-OnuI crystal structure (PDB ID 3QQY). Blue text indicates altered residues between wild type I-OnuI and eOnu_HIVInt_v1 and red text highlights the eight amino acid positions considered for variation (contacting base pairs −8, −7 and −6 of the target sequence) in the engineering of eOnu_HIVInt_v2