Figure 5.
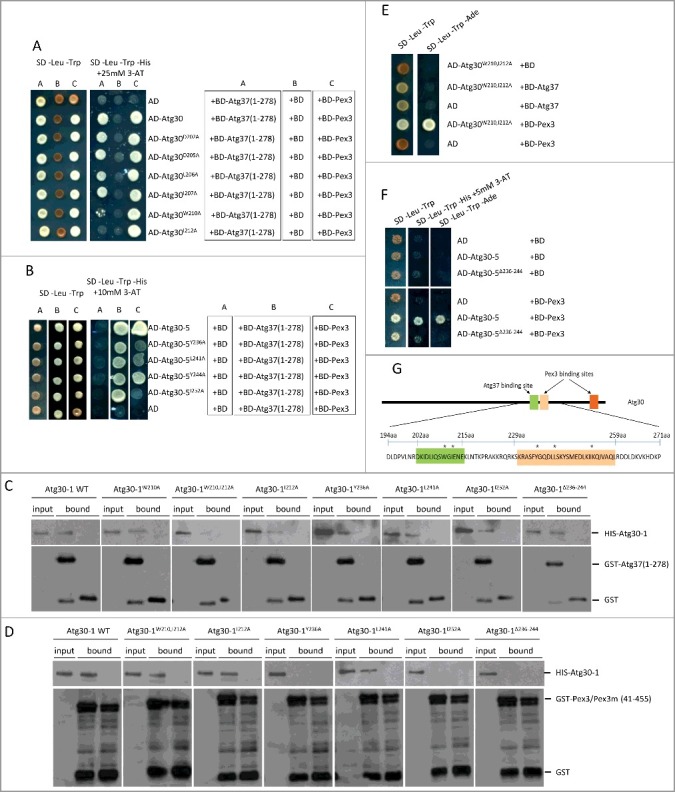
Identification of Atg37- and Pex3-binding sites in Atg30. (A) Substitutions at W210 or I212 in Atg30 affect interaction of Atg30 with Atg37, but not with Pex3. Y2H assays between AD-Atg30 WT or its mutants (Atg30D202A, Atg30D205A, Atg30L206A, Atg30I207A, Atg30W210A and Atg30I212A) and BD-Atg37(1-278) or BD-Pex3. (B) Substitutions at Y236, L241 and I252 in Atg30-5 affect the interaction of Atg30-5 with Pex3, but not with Atg37. Y2H assays between AD-Atg30-5 WT or its mutants (Atg30-5Y236A, Atg30-5L241A, Atg30-5Y244A and Atg30-5I252A) and BD-Atg37(1-278) or BD-Pex3. (C) The Atg30-1W210,I212A mutant does not bind Atg37 in vitro. Binding studies revealed that only the Atg30-1W210,I212A mutant was deficient in Atg37 binding, whereas the Atg30-1 WT and other mutants (Atg30-1W210A, Atg30-1I212A, Atg30-1Y236A, Atg30-1L241A, Atg30-1I252A and Atg30-1Δ236-244) were still pulled down with GST-Atg37(1-278). Free GST protein was used as a control. There was equivalent loading in the input and bound lanes. (D) Atg30-1Y236A, Atg30-1I252A and Atg30-1Δ236-244 mutants do not bind Pex3 in vitro. Binding studies revealed that only Atg30-1Y236A, Atg30-1I252A and Atg30-1Δ236-244 mutants were deficient in Pex3 binding, whereas Atg30-1 WT and other mutants (Atg30-1W210,212A, Atg30-1I212A, Atg30-1L241A) were still pulled down with GST-Pex3(41-455). The GST-Pex3m(41-455) protein was used as a control. There was equivalent loading in the input and bound lanes. (E) Simultaneous substitution of W210 and I212 in Atg30 strongly affects its interaction with Atg37, but not with Pex3. AD-Atg30W210,212A was used with the previously described BD-Atg37(1-278) and BD-Pex3.12 (F) Deletion of amino acids 236-244 in AD-Atg30-5 disrupts its interaction with BD-Pex3 in a Y2H assay; AD-Atg30-5 and AD-Atg30-5Δ236-244 were used with previously described BD-Pex3.12 (G) Schematic depicting the identified Atg37- (green) and Pex3- (light and dark orange) binding sites in Atg30. Asterisks indicate amino acids in the Atg30-5 region crucial for Atg37 or Pex3 binding.
