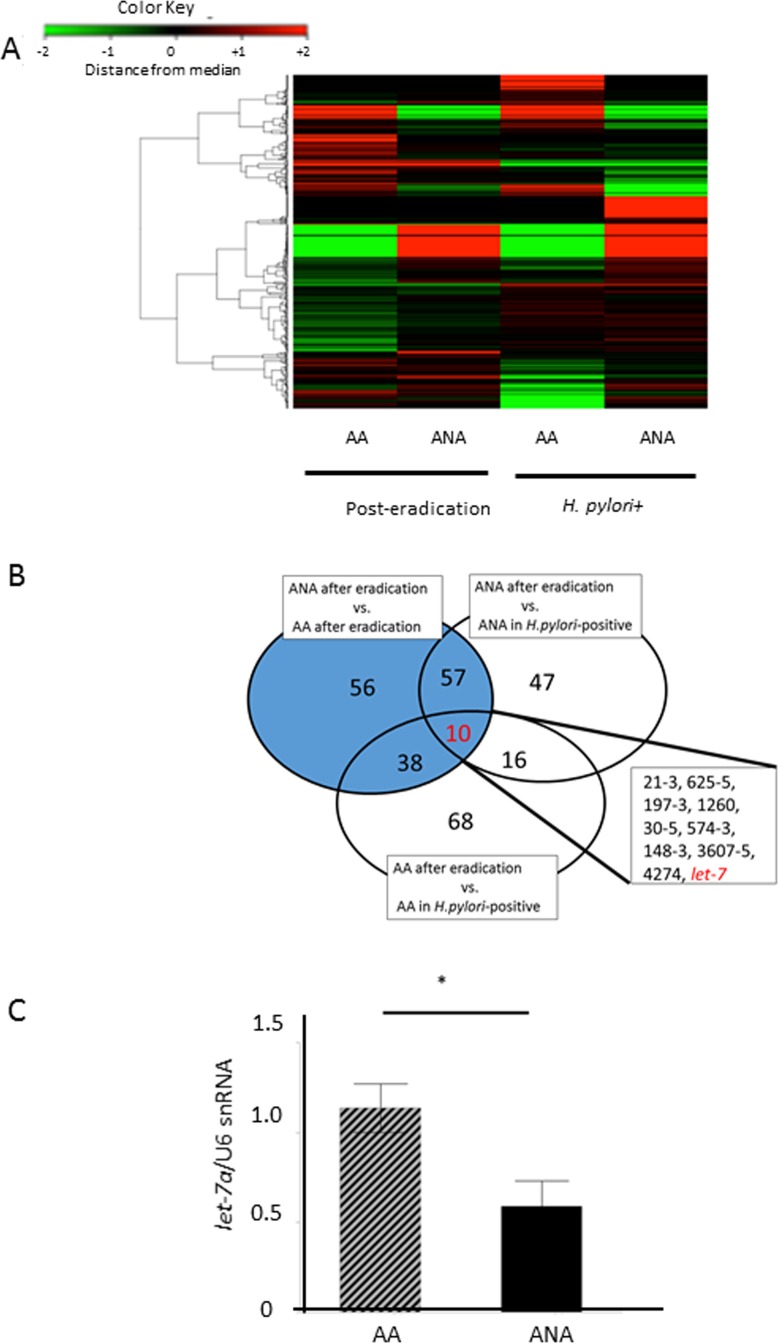
Figure 4. Identification of candidate microRNAs that cause barrier dysfunction.
(A) A heat-map of a Taqman microRNA microarray analysis in a discovery phase. There were significant differences in the microRNA profiles between samples from post-eradication mucosa and H-pylori-positive mucosa (N = 5). (B) 10 microRNAs were extracted as candidate miRNAs, including miR 21-3, miR 625-5, miR 197-3, miR 1260, miR 30-5, miR 574-3, miR 148-3, miR 3607-5, miR 4274, and let-7. Target microRNAs that yielded differences at least 2-fold or 0.5-fold expression levels were selected as candidate microRNAs., (C) In the validation phase, the expression of let-7a in ANA was significantly inhibited in comparison to AA (N = 18, P = 0.0032, Student`s t-test).