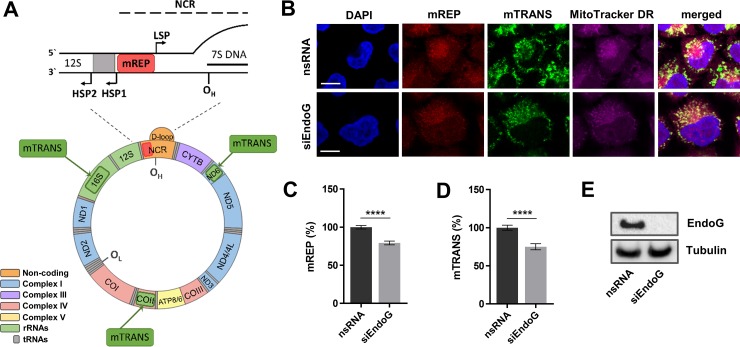
Figure 1. Assessment of EndoG's function in initiation of mitochondrial OH replication and mitochondrial transcription.
(A) Organization of the human mitochondrial genome. Recognition sites of mREP and mTRANS probes for FISH labelling with mTRIP are indicated with red and green boxes, respectively. HSP and LSP - heavy - and light-strand promoter, OH - origin of heavy strand replication, NCR - non-coding region (adopted from [7]). (B) Representative confocal microscopic images of HeLa cells stained with DAPI and MitoTracker DR (250-1000 nM, 1 h) and labelled with mREP and mTRANS probes 48 h after EndoG knockdown with siRNA. Scale bars = 10 μm. Fluorescence-based quantification of (C) mREP (initiation of mtDNA replication near OH) and (D) mTRANS (mitochondrial transcription); n = 196–218 cells from 2 independent experiments. Data are expressed as mean ± SEM (****P < 0.0001; non-parametric Mann-Whitney test for unpaired samples). (E) Western blot analysis to validate efficient knockdown of EndoG.