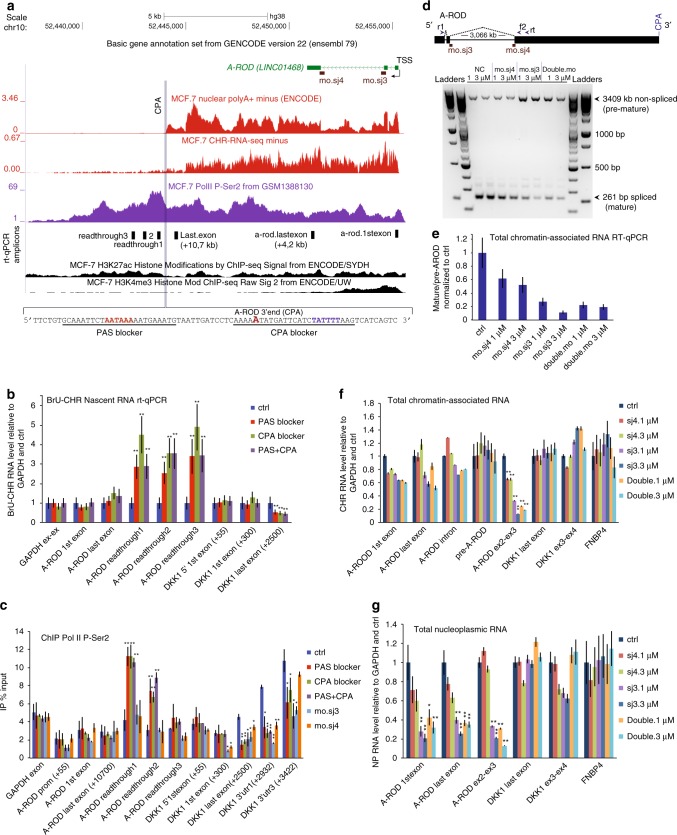
Fig. 6.
3′end formation and splicing in A-ROD function. a UCSC genomic screenshot showing A-ROD expression tracks and positions of the assayed RT-qPCR amplicons. P-Ser2 Pol II ChIP-seq is from74. Sequences of the PAS and CPA target sites of 2′-O-MeRNA-PS oligo blockers are underlined. b Normalized RT-qPCR of BrU-labeled nascent chromatin-associated RNA in MCF-7 cells treated either with control or 2′-O-MeRNA-PS oligo blockers targeting the PAS and/or CPA site. Error bars represent standard deviations from three independent experiments (n = 3 biological replicates, **p < 0.01, two-tailed Student’s t-test). c P-Ser2 Pol II ChIP recovery (% of Input) in control; MCF-7 cells treated with 2′-O-MeRNA-PS oligo blockers targeting the PAS and/or CPA site; and MCF-7 cells treated with morpholinos targeting either the donor splice site (mo.sj3) or the acceptor splice site (mo.sj4) of the second intron of A-ROD. Positions of the morpholinos are depicted in (a, d). Error bars represent standard deviations from three independent experiments (n = 3 biological replicates, *p < 0.05 and **p < 0.01, two-tailed Student’s t-test). d Schematic representation of A-ROD exonic structure and PCR analysis of the premature (non-spliced) and mature (spliced) forms of A-ROD in control MCF-7 cells (treated with the ‘standard control sequence’ morpholino) or in MCF-7 cells treated either with the mo.sj3 or the mo.sj4 in the given concentrations. In the ‘double’ lanes both morpholinos were pooled and cotransfected in the given final concentrations. The PCR (30 cycles) was performed with primers r1 and f2 on cDNA reverse-transcribed from 200 ng total nuclear RNA primed with the ‘rt’ primer (see Methods section). Please note, that this PCR analysis is not quantitative and was conducted to visualize the effect of the morpholinos treatment. The accumulation of the premature product observed in lanes 5–8 (mo.sj3 and double.mo) is most probably due to the mo.sj3-exerted elimination of the mature form of A-ROD (hence less available spliced product to amplify) and both products being amplified by the same pair of primers, in the same reaction. To quantitatively assess the morpholino treatment effect, RT-qPCR analysis was further performed (e–g). e Normalized to control RT-qPCR measured abundance of the mature (spliced) form of A-ROD (amplicon ‘exon2–exon3’) divided to the RT-qPCR measured level of the premature (non-spliced) form (amplicon ‘pre-A-ROD.2’), in control and in MCF-7 cells treated with morpholinos as in (d). The RT-qPCR values used to extract the ratio spliced/pre-mature A-ROD are from RT-qPCR on total chromatin-associated RNA (cDNA primed with random primers) depicted in (f). f, g RT-qPCR analysis on f total chromatin-associated RNA and g nucleoplasmic RNA (in both cases random primed cDNA) in control and MCF-7 cells treated with morpholinos as in (d). Error bars represent standard deviations from three independent experiments (n = 3 biological replicates, *p < 0.05 and **p < 0.01, two-tailed Student’s t-test)