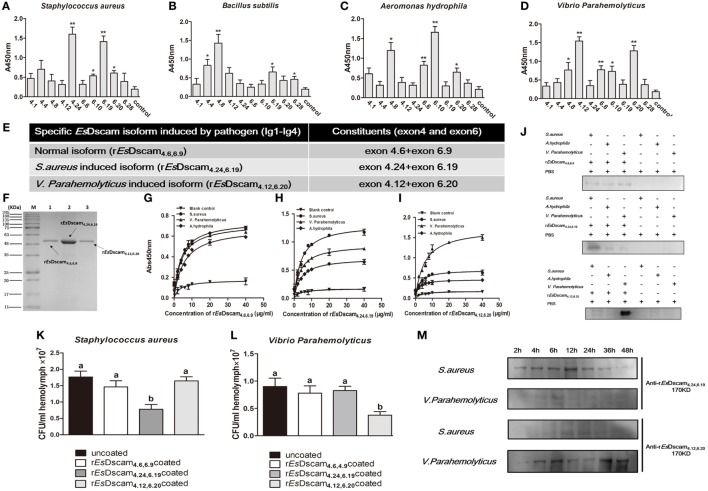
Figure 4.
Soluble EsDscam-specific binding with bacteria and promotes its clearance. (A–D) Bacterial binding specificity of epitope II in exon 4/exon 6. The binding activity between selected peptides and different bacteria were analyzed by ELISA with peptides that containing the conserved motif as control. Three independent repeats were performed, and results are expressed as the mean ± SD. Data were analyzed by Student’s t-test. *p < 0.05, **p < 0.01. (E) Specific recombinant EsDscam isoforms induced by pathogens. Normal isoform composed of alternative spliced exon 4.6, alternative spliced exon 6.9, and constant exon designated rEsDscam4.6,6.9; Staphylococcus aureus-induced isoform composed of alternative spliced exon 4.24, alternative spliced exon 6.19, and constant exons designated rEsDscam4.24,6.19; Vibrio parahemolyticus-induced isoform composed of alternative spliced exon 4.12, alternative spliced exon 6.20, and constant exons designated rEsDscam4.12,6.20. (F) Purified recombinant specific EsDscam isoform. Three recombinant rEsDscam protein were expressed from the pET-28a (+) vector in Escherichia coli Rosetta (DE) cells and purified by affinity chromatography, followed by SDS-PAGE detection. (G–I) Quantitative binding of EsDscam with bacteria. Microtiter plates were coated with heat-inactivated bacteria which were pre-coated with serial concentrations of proteins and air dried at 37°C for 2 h. After the wells were blocked with 2% BSA, the bound proteins were then detected by ELISA using an anti-His-tag Ab. Data represent the mean ± SD of three independent experiments. (J) Microorganism binding assays of rEsDscam proteins. The coated samples of bacteria with or without rEsDscam protein and PBS were boiled for 10 min and separated by 12% SDS-PAGE for Western blotting using an anti-His-tag antibody, with PBS incubated with microorganisms as control. Data are representative of two independent repeats. (K,L) EsDscam promotes bacteria clearance in crab. Each rEsDscam (5 µg) was incubated with 100 µl of the S. aureus (left panel) or V. parahemolyticus (right panel) suspension (OD600 = 0.2) for 1 h. The bacteria were washed and suspended in 200 µl of PBS. Then, 200 µl of the suspension was injected into the crab, and hemolymph was collected 30 min later. The number of residual bacteria in the hemolymph was determined by plating onto LB agar plates. Uncoated bacteria were used as the control. Three to five crabs were used for each group. Three independent repeats were performed, and results are expressed as the mean ± SD. Data were analyzed by one-way ANOVA and the letters (a, b) presented significant differences (p < 0.05). (M) Specific EsDscam was highly induced after bacteria stimulation in cell-free hemolymph. Protein from cell-free hemolymph was extracted at each time point after bacterial stimulation and analyzed by Western blot using anti-rEsDscam4.24,6.19 and anti-rEsDscam4.12,6.20 antibodies. Each sample was from four crab. Data are representative of two independent repeats.