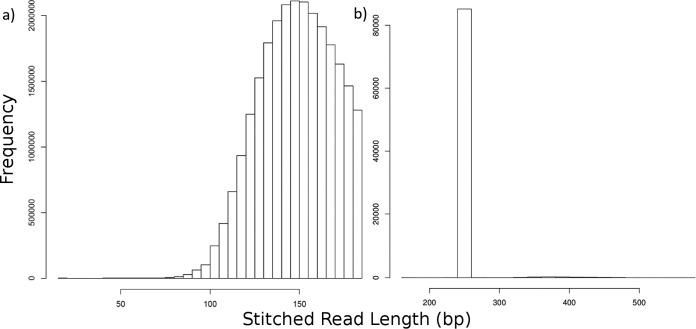
FIG 2 .
Histogram of stitched read lengths in a single Human Microbiome Project (HMP) metagenomic sample (a) and 16S V4 Primate Microbiome Project (PMP) sample (b). (a) A shotgun metagenomic sample produces stitched contigs spanning a range of lengths. The truncation after read lengths of 185 bp is due to enforcing a minimum overlap length of 15 base pairs, which in a data set consisting of 100-bp reads is the maximum allowable length (100 + 100 − 15). Because the mean of this distribution is 148.6 and its standard deviation is 20.62, the coefficient of variation (CV) is 0.139, above the 0.1 threshold under which the data would be considered amplicon-like by default; the data are hence considered shotgun reads by SHI7. (b) A 16S amplicon sample produces a distinct histogram marked by high representation of certain contig lengths corresponding to target gene size, in this case 252 and 253 base pairs, and a much lower CV (mean = 254.4, SD = 15.7; CV = 0.062). Most residual longer reads match PhiX174, an Illumina control contaminant, and are later removed by SHI7 in “learning mode” by filtering out sequences within a mean read length ± SD/2 in amplicon samples.