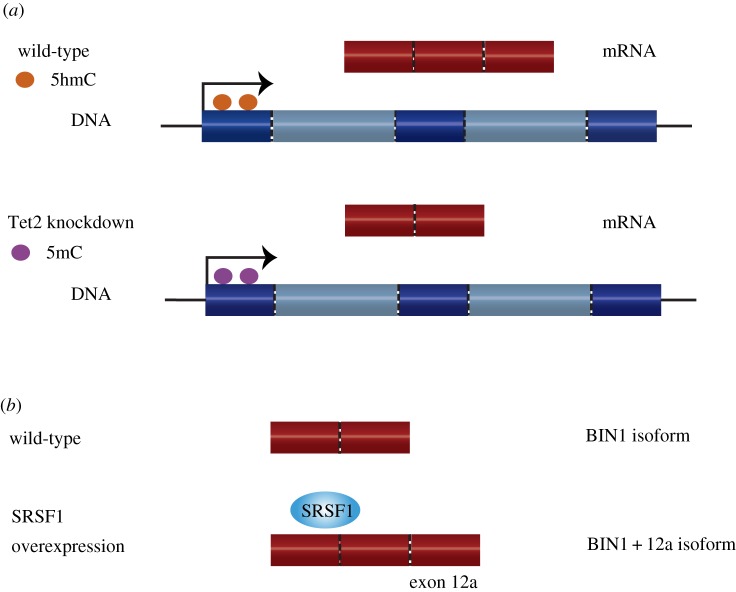
Figure 1.
(a) Example of epigenetic markers influencing alternative splicing. Knockdown of the oxidase Tet2 has been shown to reduce levels of cytosine hydroxymethylation (5hmC) (orange circles). For some genomic regions with a reduction in 5hmC, a corresponding increase in cytosine methylation (5mC) was observed (blue circles). 5hmC was shown to be prevalent in exons, particularly at the boundaries, and the loss of 5hmC resulted in differential inclusion of exons. (b) The overexpression of splicing factor SRSF1 is known to cause differential splicing of BIN1. Overexpression of SRSF1 results in an alternative isoform BIN1+12a. This isoform is unable to bind the tumour repressor cMYC and therefore promotes tumorigenesis.