Figure 3.
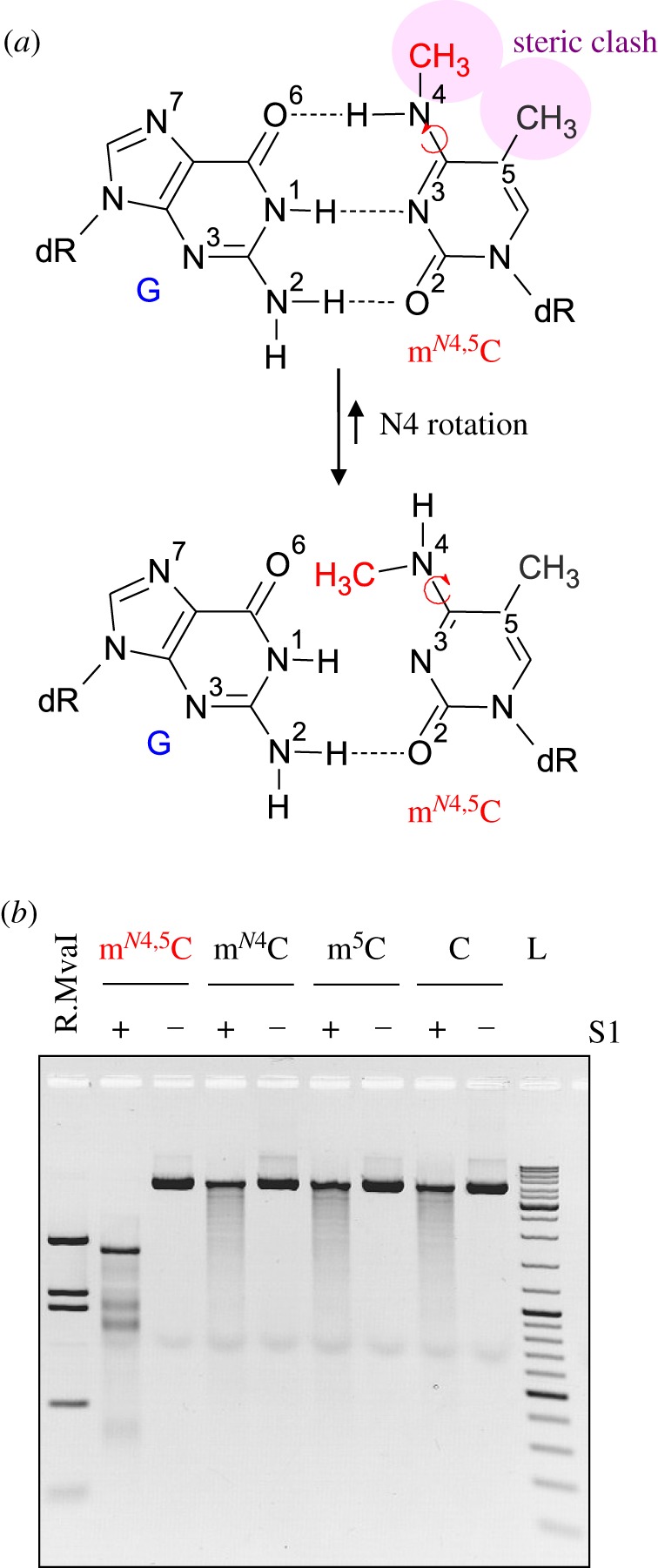
The mN4,5C residue disrupts normal DNA structure. (a) Structural considerations of the mN4,5C residue in dsDNA [20,36,37] show how rotation around the N4–C4 bond of mN4,5C relieves the steric clash between the methyl groups (upper panel) but at the same time disrupts the normal G · C hydrogen-bonding pattern (lower panel), precluding a normal Watson–Crick DNA structure. (b) Impact of mN4,5C on DNA structure probed by susceptibility to nuclease S1 digestion. pBR322 plasmid DNA containing mN4,5C, mono-methylated (either mN4C or m5C) or unmethylated cytosine (c), respectively, at CC(A/T)GG sites (electronic supplementary material, figure S9) was incubated with nuclease S1 and analysed by agarose gel electrophoresis. R.MvaI lane, commercial pBR322 digested with R.MvaI; L, GeneRuler DNA Ladder Mix.
