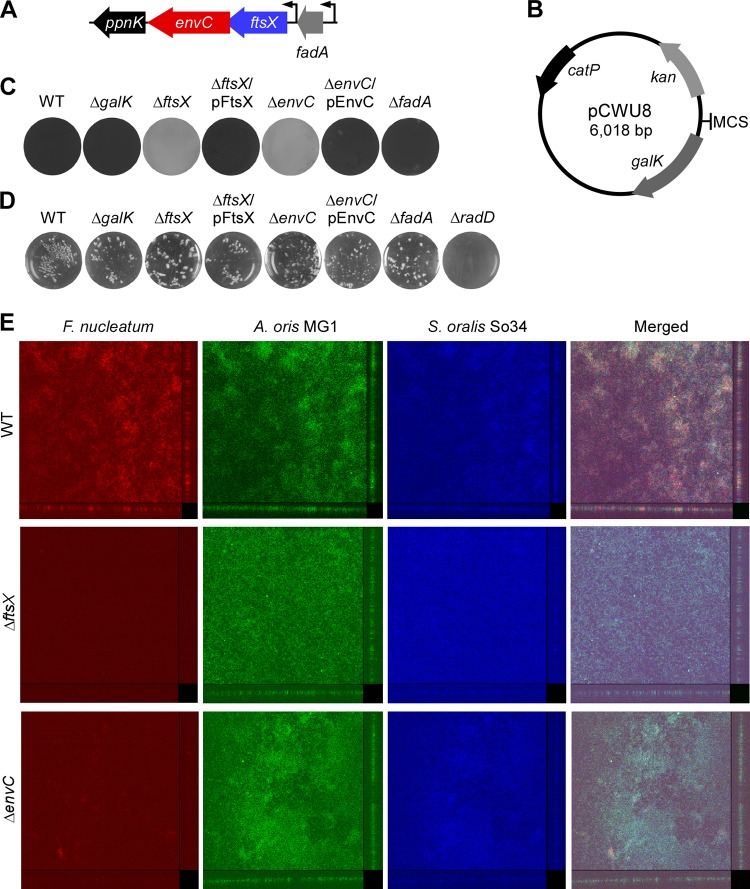
FIG 1 .
Roles of fusobacterial ftsX and envC in the development of monospecies and multispecies biofilms. (A) ftsX locus in the chromosome of F. nucleatum ATCC 23726, with envC adjacent to ppnK, which encodes an NAD+ kinase. Upstream of ftsX is fadA coding for the adhesin FadA (6); expression of genes in the ftsX locus and fadA appears to be controlled by individual promoters. (B) Map of the nonreplicative vector pCWU8 used to generate unmarked, in-frame gene deletion mutants in F. nucleatum. The kanamycin resistance cassette (kan), chloramphenicol/thiamphenicol resistance gene (catP), and galK are indicated. The multiple cloning sites (MCS) contain EcoRI, SacI, KpnI, BamHI, and SalI. (C) Fusobacterial strains were evaluated for their ability to form monospecies biofilm using crystal violet staining. (D) Interaction of fusobacteria and S. oralis So34 was determined by a standard coaggregation assay, with a radD mutant used as a negative control. (E) Three-species biofilms were analyzed by confocal laser scanning microscopy at a magnification of ×20, using 16S rRNA-oligonucleotide probes specific for F. nucleatum (red), A. oris (green), and S. oralis (blue); side and top views are presented. The results are representative of three independent experiments performed in triplicate.