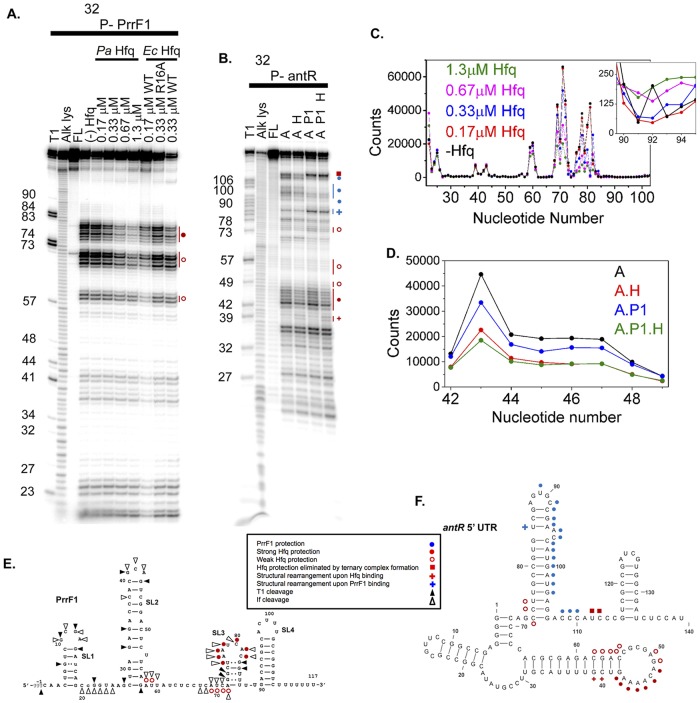
FIG 4.
RNase If footprinting of PrrF1 and antR. (A) RNase If digestion pattern of 5′-32P-PrrF1 sRNA at 30°C with no Hfq [(−) Hfq], 0.17 to 0.67 μM P. aeruginosa (Pa) Hfq, 0.17 to 0.33 μM E. coli (Ec) Hfq, and 0.33 μM E. coli R16A Hfq, as shown. Lanes T1, Alk lys, and FL represent control reactions in urea with RNase T1 (T1), alkaline hydrolysis (Alk lys), and no treatment (FL). E. coli Hfq binds sRNA more strongly than P. aeruginosa Hfq, and binding of a second E. coli Hfq hexamer may account for some differences between the protection pattern in 0.17 and 0.33 μM E. coli Hfq. (B) RNase If digestion patterns of 5′-32P-labeled antR mRNA at 30°C with no Hfq (lane A), 0.5 μM P. aeruginosa Hfq (lane A H), 0.5 μM PrrF1 (lane A P1), and 0.5 μM P. aeruginosa Hfq plus 0.5 μM PrrF1 (lane A P1 H). Control reactions were as for panel A. Numbers to the left of the images in panels A and B indicate the nucleotide numbers of the RNA molecule where cleavage occurs to produce the observed bands. (C) Relative digestion of PrrF1 sRNA by RNase If in the absence and presence of P. aeruginosa Hfq (see Materials and Methods). The inset shows the region between nt 90 and 95. (D) Relative digestion of antR mRNA by RNase If in the absence and presence of P. aeruginosa Hfq and PrrF1 sRNA. (E and F) Summary of nuclease digestion patterns on the predicted secondary structures of PrrF1 (E) and antR 5′ UTR (F), as indicated in the key. The schematics summarize the results of several footprinting experiments for each RNA. Weak (<2×) and strong (∼2×) protection were determined by quantitation of the footprinting gels with SAFA (71), as illustrated in panels C and D. A plus sign indicates increased nuclease cleavage upon Hfq (red) or PrrF1 (blue) binding, suggesting structural rearrangements of the RNA molecules. The secondary structures of the PrrF1 sRNA and antR mRNA shown in panels E and F, respectively, were generated using a combination of computational prediction (MFold) (73) and experimental results (A and B).