Figure 1.
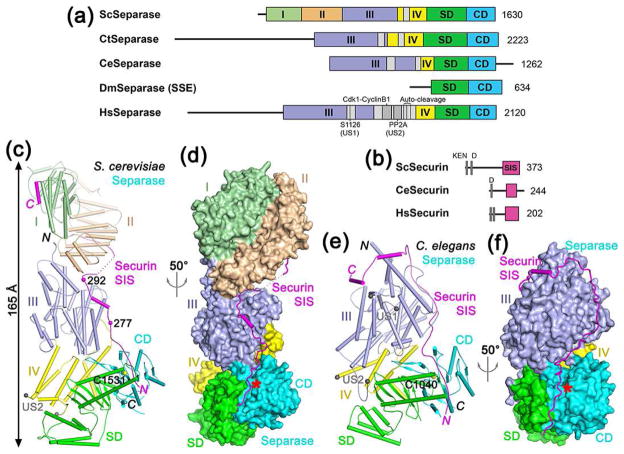
Structures of separase-securin complexes. (a). Domain organization of separase. The helical region of yeast separase is divided into four domains (I–IV) and given different colors, which is followed by SD (substrate-binding domain) and CD (catalytic domain). The domain boundaries for the N-terminal region of C. thermophilum and human separase are not known and therefore are not indicated. The unstructured segments (US) in the helical region are shown in gray. For Drosophila separase, only the subunit containing the SD-CD (known as SSE) is shown. Sc: S. cerevisiae (yeast), Ct: Chaetomium thermophilum, Ce: C. elegans, Dm: D. melanogaster, Hs: Homo sapiens. (b). Domain organization of securin. The separase interaction segment (SIS) is shown in magenta. The N-terminal KEN and D-boxes are indicated. (c). Schematic drawing of the structure of the yeast separase-securin complex. The domains of separase are colored as in Fig. 1a, and the securin SIS is in magenta. The catalytic Cys1531 is shown as a sphere model. Two of the phosphorylation sites in securin are indicated with spheres. The ends of the unstructured segment (US2) are indicated by the two gray spheres. (d). Structure of the yeast separase-securin complex, with separase shown as a molecular surface, viewed after 50° rotation around the vertical axis from panel c. The active site of separase is indicated with the red star. (e). Schematic drawing of the structure of the C. elegans separase-securin complex. The catalytic Cys1040 is shown as a sphere model. (f). Structure of the C. elegans separase-securin complex, with separase shown as a molecular surface. All structure figures were produced with PyMOL (www.pymol.org).