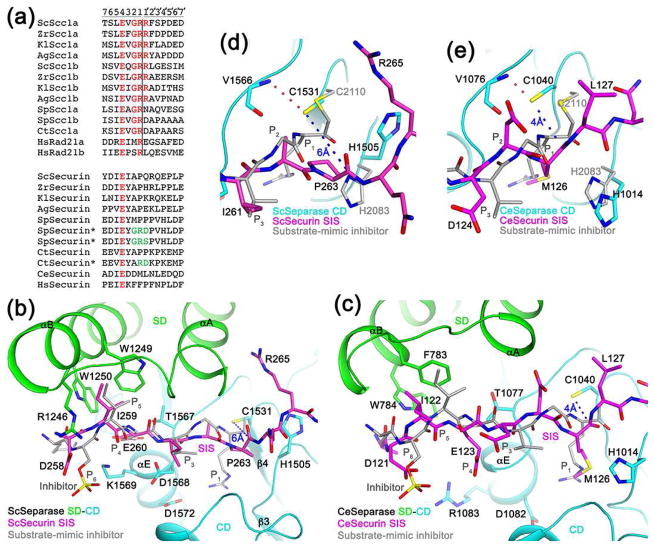
Figure 3.
Molecular mechanism for the inhibition of separase by securin. (a). Alignment of the cleavage sites in separase substrates. The two cleavage sites in each protein are named a and b. The equivalent residues in securin are also shown. The asterisks indicate securin mutants (mutations in green) that become substrates of separase. The P and P′ residues are labeled at the top, and the cleavage site is indicated with the vertical line. (b). Binding mode of residues 258-265 of yeast securin SIS (magenta) in the active site of yeast separase. Side chains of residues in the interface are shown as stick models and labeled. The bound position of a substrate-mimic inhibitor to C. thermophilum separase SD-CD is shown in gray. (c). Binding mode of residues 121-127 of C. elegans securin SIS (magenta) in the active site of C. elegans separase. (d). Closeup of the active site region of yeast separase. The catalytic Cys1531 side chain is hydrogen-bonded to the main-chain amide of Val1566 (dashed line in red) and 6 Å from the carbonyl carbon of Pro263 (dashed line in blue). (e). Close-up of the active site region of C. elegans separase.