Figure 3.
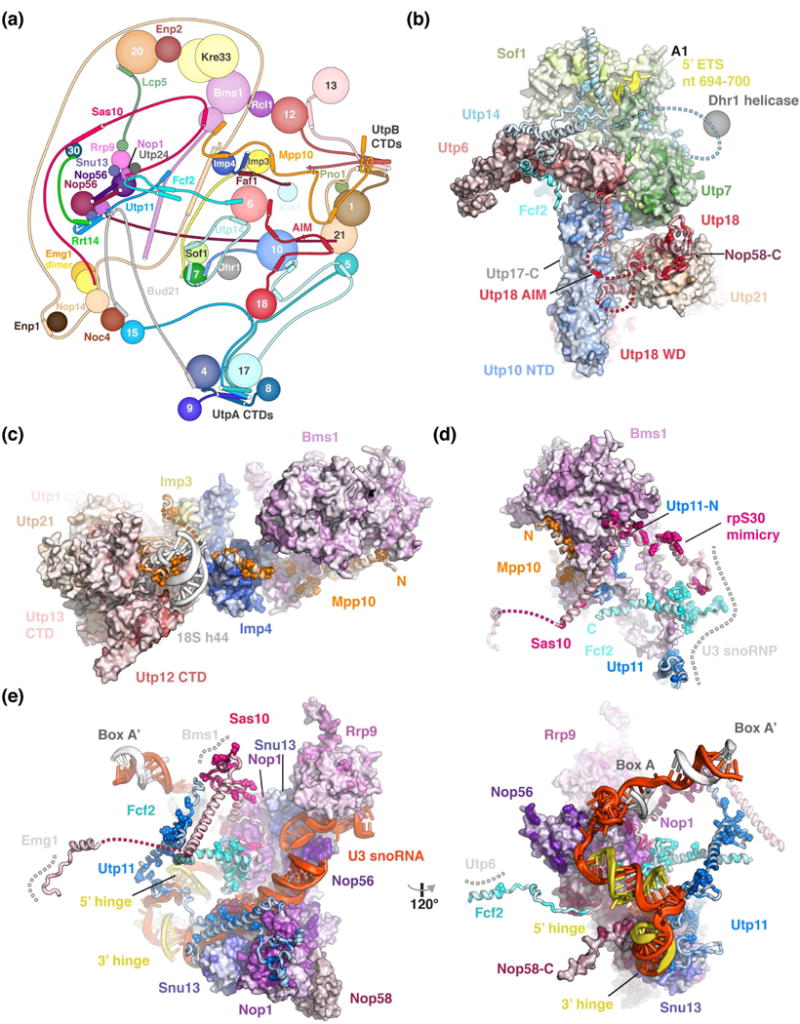
Peptides connect distant sites within the SSU processome via conserved binding motifs. (a) Schematic protein-protein interaction diagram of selected SSU processome components represented as spheres or lines with interacting elements as helices or strands. The Utps (U-three proteins) are labeled with their respective number. (b-e) Detailed views of Utp18 and Utp14 (b), Mpp10 (c), Bms1 (d) and the U3 snoRNP (e) with proteins shown as surface or cartoon, colored according to conservation with residues conserved more than 90 % highlighted as spheres. Direct interaction partners depicted as surface or a grey dashed line. All proteins are colored by conservation from lighter to darker shades. Clustal [49] was used to align manually curated sequences (H. sapiens, S. cerevisiae, G. gallus, D. melanogaster, S. pombe, C. elegans, D. rerio, A. thaliana, A. gambiae, P. troglodytes, R. norvegicus, M. musculus, B. taurus, S. scrofa) and plotted onto the structure using Homolmapper [50].
