Figure 4.
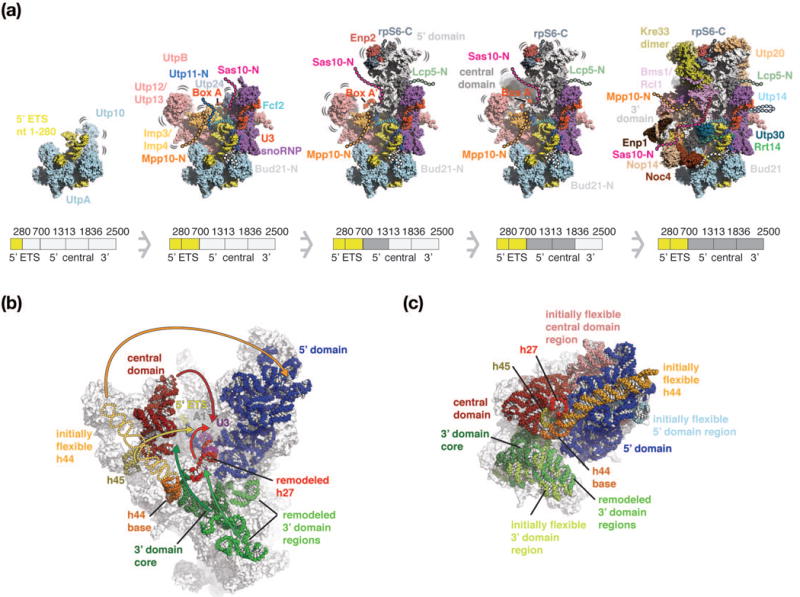
Three-dimensional maturation model of the Saccharomyces cerevisiae small-subunit processome. (a) Co-transcriptional assembly of the SSU processome as a function of transcription of rRNA regions (280 nucleotides of 5′ ETS, 5′ ETS, 5′ domain, central domain and 3′ domain). 5′ ETS (yellow), rRNA domains (white) and complexes such as UtpA (light-blue), UtpB (red), U3 snoRNP (purple, red), the Mpp10 complex (orange) and additional proteins, are shown as surfaces. Bound peptides are shown as cartoon with their initially flexible tails as dashed lines. As maturation progresses, these tails recruit additional factors and become ordered. The pre-rRNA shown in the intermediates is schematically indicated below each particle and colored in darker shades. As this model is based on the mature SSU processome structure, regions that are presumed to be flexible in earlier states are highlighted accordingly. (b, c) Comparative locations of rRNA domains within the SSU processome [PDB 5wlc] (b) and the mature small ribosomal subunit [pdb 4v88] (c). Individual rRNA domains are colored identically with 5′ domain (blue), central domain (red), 3′ domain (green) and shown as spheres superimposed onto transparent outlines of the particles. In the SSU processome, the flexible helix 44 is indicated as schematic outline. Rearrangements of rRNA domains from the SSU processome (b) that are necessary to obtain the positions within the mature small ribosomal subunit (c) are indicated with arrows. The central U3 snoRNA Box A and Box A′ are colored in purple. RNA elements disordered in the SSU processome are indicated in lighter shades in the mature SSU.
