Abstract
Patient-Derived Xenografts (PDXs), entailing implantation of cancer specimens in immunocompromised mice, are emerging as a valuable translational model that could help validate biologically relevant targets and assist the clinical development of novel therapeutic strategies for gastric cancer.
More than 30% of PDXs generated from gastric carcinoma samples developed human B-cell lymphomas instead of gastric cancer. These lymphomas were monoclonal, Epstein Barr Virus (EBV) positive, originated tumorigenic cell cultures and displayed a mutational burden and an expression profile distinct from gastric adenocarcinomas. The ability of grafted samples to develop lymphomas did not correlate with patient outcome, nor with the histotype, the lymphocyte infiltration level, or the EBV status of the original gastric tumor, impeding from foreseeing lymphoma onset. Interestingly, lymphoma development was significantly more frequent when primary rather than metastatic samples were grafted.
Notably, the development of such lympho-proliferative disease could be prevented by a short rituximab treatment upon mice implant, without negatively affecting gastric carcinoma engraftment.
Due to the high frequency of human lymphoma onset, our data show that a careful histologic analysis is mandatory when generating gastric cancer PDXs. Such care would avoid misleading results that could occur if testing of putative gastric cancer therapies is performed in lymphoma PDXs. We propose rituximab treatment of mice to prevent lymphoma development in PDX models, averting the loss of human-derived samples.
Introduction
Recent clinical experience suggests that several prerequisites are mandatory for the success of a targeted therapy: i) a precise molecular annotation of tumors, to identify possible key targets; ii) the proper selection of patients that could benefit from that therapy; iii) the validation of the true biological relevance of these targets in the context of a specific tumor type; iv) the validation of the inhibitor(s) of the identified target(s) that is/are more efficient in that particular context; v) the identification of molecular alterations predicting responsiveness to treatment.
At the moment, the best strategy to fulfill all of the above mentioned prerequisites is the use of Patient-Derived Xenografts (PDXs), entailing subcutaneous implantation of fresh cancer samples in immunocompromised mice, subsequently expanded to generate a molecularly annotated study population that can be randomized for prospective treatment with molecular therapies.[1], [2]
In patients affected by gastric tumors, the use of targeted therapies is much less common than in other cancer patients, with only two molecular drugs approved so far, namely Trastuzumab and Ramucirumab.[3], [4], [5] Pre-clinical models such as PDXs could help validate biologically relevant targets and assist the clinical development of novel combination strategies for this disease.6
PDXs are generated in mice lacking an active immune system that show a relatively high frequency of lymphoproliferative diseases and are vulnerable to T-cell-controlled infections, in particular to the Epstein Barr Virus (EBV).[7], [8] This virus (affecting around 90% of humans) can infect both immune (human B-cells, T-cells, NK cells) and epithelial cells,9 where it persists for life.10 In humans, EBV is responsible not only for infectious mononucleosis but it is also associated with malignancies, such as nasopharyngeal and gastric carcinoma, and lymphoproliferative diseases, like Hodgkin lymphoma, Burkitt lymphoma, and lymphomas in immune-depressed patients.10 Here, we report that more than 30% of PDXs generated from gastric tumor samples developed EBV-positive lymphomas of human origin instead of gastric carcinomas. Interestingly, the ability of grafted samples to develop lymphomas did not correlate with the amount of lymphocytic infiltration of the original tumor or with EBV positivity, while it was significantly higher when primary rather than metastatic samples were grafted. Importantly, the development of these lympho-proliferative diseases could be prevented by mice treatment with rituximab upon grafting, thus avoiding the loss of human-derived adenocarcinoma samples.
Materials and Methods
Specimen Collection
Gastric tumor samples and matched normal samples were obtained from patients undergoing surgery in 12 Italian Hospitals (Supplementary Materials and Methods). All patients provided informed consent; samples were collected and the study was conducted under the approval of the Review Boards of all the Institutions. Clinical and pathologic data were entered and maintained in our prospective database. All the samples have been anonymized before being shipped to Candiolo. No references to the patients can be inferred from the immunohistochemical and molecular characterization presented in the work.
PDX Generation
Gastric cancer material from Torino Hospitals was collected in medium 199 supplemented with 100 μg/mL levofloxacin, stored at 4°C and received within few hours from surgical resection. Gastric cancer material from other Italian Hospitals was collected in IGL-1® preservation medium supplemented with 100 μg/mL levofloxacin, stored at 4°C and received within 24 hours from surgical resection. At Candiolo Cancer Institute each sample was cut into 25- to 50-mm3 pieces in antibiotic-containing 199 medium; some of the pieces were incubated overnight in RNAlater and then frozen at −80°C for molecular analyses; one piece was fixed in Formalin; 2 pieces were coated in Matrigel (BD Biosciences) and implanted in two/three different 4/6-week-old female NOD/SCID mice. After mass formation, tumors were passaged and expanded for at least 2 generations. Tumor size was evaluated by caliper measurements and the approximate mass volume was calculated using the formula 4/3π.(d/2)2.D/2, where d is the minor tumor axis and D is the major tumor axis. The genetic identity between the original tumor and the corresponding PDX was evaluated by short tandem repeat profiling (Cell ID, Promega, Madison, WI, USA). CRC PDXs were established as described in.11 If not differently specified in the text, the different analyses have always been performed on first passage PDXs. Tumor growth rate was calculated using the formula (Vf-Vi)/T, where Vf is the final volume, Vi is the initial volume, T is the time (number of days from the initial and the final measurements).
All animal procedures were approved by the Ethical Commission of the Candiolo Cancer Institute (Candiolo, Torino, Italy), and by the Italian Ministry of Health.
Analyte Extraction
Genomic DNA was isolated using the Relia Prep gDNA Tissue Miniprep System (Promega). Total RNA was extracted using the miRNeasy Mini Kit (Qiagen) and quality checked by measuring the 28S/18S ribosomal RNA ratio with an Agilent 2100 Bioanalyzer (Agilent Technologies). DNA and RNA concentrations were quantified using a Nanodrop 1000 Spectrophotometer (Thermo Fisher Scientific).
Microarray Data Generation
Synthesis of cDNA and biotinylated cRNA (from 500 ng total RNA) was performed using the IlluminaTotalPrep RNA Amplification Kit (Ambion), according to the manufacturer’s protocol. Quality assessment and quantitation of cRNAs were performed with Agilent RNA kits on a Bioanalyzer 2100 (Agilent). Hybridization of cRNAs (750 ng) was carried out using Illumina Human 48k gene chips (Human HT-12 V4 BeadChip). Array washing was performed by Illumina High Temp Wash Buffer for 10′ at 55°C, followed by staining using streptavidin-Cy3 dyes (Amersham Biosciences). Hybridized arrays were stained and scanned in a Beadstation 500 (Illumina).
Microarray Data Preprocessing and Statistical Analysis
Probe intensity data were extracted using the Illumina Genome Studio software (Genome Studio V2011.1). Probes were filtered to select those showing detectable signal (detection P value = 0) in at least one sample.
Hierarchical clustering of the samples was performed on Euclidean distance with pvclust package12 in R bioconductor[13], [14] considering the top 1000 most variables probes.
Analysis of IG Heavy Chain Rearrangement
Rearrangements in the immunoglobulin heavy chain gene (IgH) were analyzed using the IgH Rearrangements Molecular Analysis Kit (Master Diagnòstica, Spain) according to the manufacturer’s instructions. This kit allows detection of clonality in B cell lymphoproliferative processes by PCR amplification of rearranged VDJ segments (FR1-JH, FR2-JH and FR3-JH) in the hypervariable region of IgH, using multiple primers that hybridize with conserved regions in the genes. The DNA quality was checked for all samples using the p53 control gene. PCR products were evaluated by heteroduplex analysis and electrophoresis in polyacrylamide gels (Criterion precast gel, BIO-RAD), stained with SYBR Safe (Thermo Fisher Scientific) and visualized under UV light. When DNA is amplified from a clonal lymphoid population, the resulting fragments are all identical in size and sequence, giving a single band.
EBV Evaluation
The detection and quantification of EBV DNA were performed using the EBV Q-PCR Alert KIT (ELITechGroup S.p.A., Italy). The real-time amplification assay was carried out on ABI 7300 Real-Time PCR System instrument (Applied Biosystems, USA). EBV ISH was performed using the Bond™ Ready-to-Use ISH EBER Probe and the L Bond™ Ready-to-Use Anti-Fluorescein Antibody (Leica Biosystems, Newcastle Ltd, United Kingdom).
Rituximab Treatment
Animals treated with Rituximab received the drug 4 days after the subcutaneous (s.c.) implant of the tumor mass, with the following regimens: (i) Rituximab i.p. 25 mg/kg, 3 times/week for 1 week; (ii) Rituximab i.p. 25 mg/kg, 3 times/week for 1 week and once a week for the following 3 weeks; (iii) Rituximab i.v. 10 mg/kg, 3 times/week for 1 week. Rituximab was provided by the Hospital Pharmacy that collected small amount of drugs left over after treating patients.
Immunohistochemistry
Immunohistochemistry was performed on FFPE tissue sections using an automated slide instrumentation platform DAKO and the following primary antibodies: pan-Cytokeratin, CD3, CD20, CD30, CD45, Bcl-2, Bcl-6, ALK1 (DAKO, Carpinteria, CA); CD5, CD10 (Novocastra, Newcastle upon Tyne, England); Cyclin D1 (Thermo Fischer Scientific, Waltham, MA, USA). Antibody dilutions are reported in Supplementary Table 1. Deparaffinization, rehydration, and target retrieval were performed in the PT Link (Dako PT100). Slides were then processed on the Autostainer Link 48 (Dako AS480) using an automated EnVision FLEX (DAKO) staining protocol. Positive and negative controls were included for each immunohistochemical run. Pictures were acquired with the Leica Assistant Suit (LAS EZ) Software. Uneven illumination was corrected using a control image as described in.15
Lymphoma Cell Lines Culturing
Lymphoma cells were derived from lymphoma PDXs as described in16 for Gastric PDX-derived cells.
Cytogenetic and Fluorescence In Situ Hybridization (FISH) Analysis
Cytogenetic analysis was performed on the PDX lymphoma cells. Ten G-banded metaphases were analyzed, and the karyotype was described according to the International System for Human Cytogenetic Nomenclature.17 FISH analysis was performed to evaluate the BCL2, MALT2 and IGH gene status. For this purpose, the following probes were used: ZytoLight® SPEC BCL2 Dual Color Break Apart Probe (ZytoVision GmbH, Germany), ZytoLight® SPEC MALT1 Dual Color Break Apart Probe (ZytoVision GmbH) and IGH Breakapart Probe (Cytocell, Cambridge UK). One hundred nuclei were analyzed.
Genomic Sequencing
DNA extracted from PDX models along with a sample of normal germline DNA from each patient were utilized for next generation sequencing. Using standard methods, Illumina sequencing libraries were generated and subjected to hybrid capture with a focused targeted bait set of 243 genes selected based upon their alteration in prior studies of gastroesophageal cancer. Somatic events were identified with MuTect v1.1.418 and Indelocator (http://www.broadinstitute.org/cancer/cga/indelocator). Additionally, sequencing was performed on a germline DNA sample from NOD-SCID mice and putative somatic alterations detected in PDXs that were also identified in murine DNA were filtered and removed.
Statistics
Statistical Analysis
Statistical testing was performed with GraphPAD PRISM Software 7.02, using the test indicated in Figure Legends. Statistical significance: ns = not significant; *P < .05; **P < .01; ***P < .001; ****P < .0001;.
Results
Onset of Human B Cell Lymphomas Upon Implant of Gastric Carcinomas
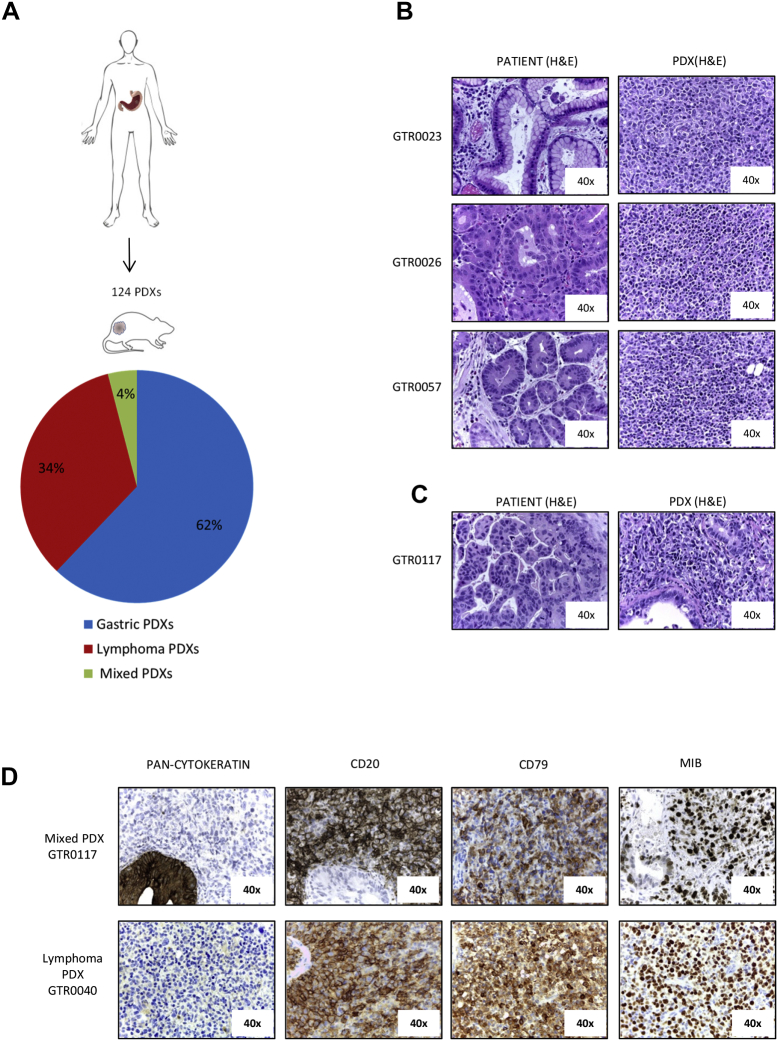
We established a platform of 124 PDXs in NOD-SCID mice by subcutaneous transplantation of fresh surgical gastric tumor samples (Figure 1A). All the tumors grown in mice were histologically evaluated; around 34% of them, at the first generation in mice, did not resemble the original tumor but rather displayed features of lymphomas (Figure 1, A and B). Histologically, most were classified as large cell lymphomas, being populated by mononuclear cells, ranging from small lymphocytes or plasma cells to large atypical lymphoid cells with pleomorphic nuclei and abundant basophilic cytoplasm, resembling lymphoblastoid cells (Figure 1B). In 4% of cases, we detected foci of carcinoma cells dispersed among non-epithelial cells (subsequently referred as ‘Mixed PDXs’ (Figure 1, A, C and D).
Figure 1.
B-cell lymphomas originate from gastro-esophageal adenocarcinomas implanted in NOD/SCID mice. A. Fresh surgical gastric adenocarcinoma samples were subcutaneously implanted in NOD/SCID mice. The pathological evaluation of 124 established first generation PDXs revealed that 77 were gastric adenocarcinomas (62%), 42 were lymphomas (34%) and 5 were mixed tumors, presenting both gastric and lymphoma tissues in the same tumor mass (4%). B. Haematoxylin and Eosin (H&E) staining of three representative primary gastric tumors (left panels) and the derived PDXs, which were diagnosed as large cell lymphomas (right panels). C. H&E staining of a primary gastric tumor (left panel) and the derived mixed PDX (right panel), showing epithelial gastric tumor cells interspersed among lymphoma cells. D. Representative immunohistochemical analysis of one lymphoma PDX and of one mixed PDX derived from primary gastric cancers. Pan-cytokeratin, CD20, CD79 and MIB (KI-67) staining were performed. As shown, lymphoma cells were pan-cytokeratin negative, CD20 and CD79 positive, and exhibited a high MIB-positive score.
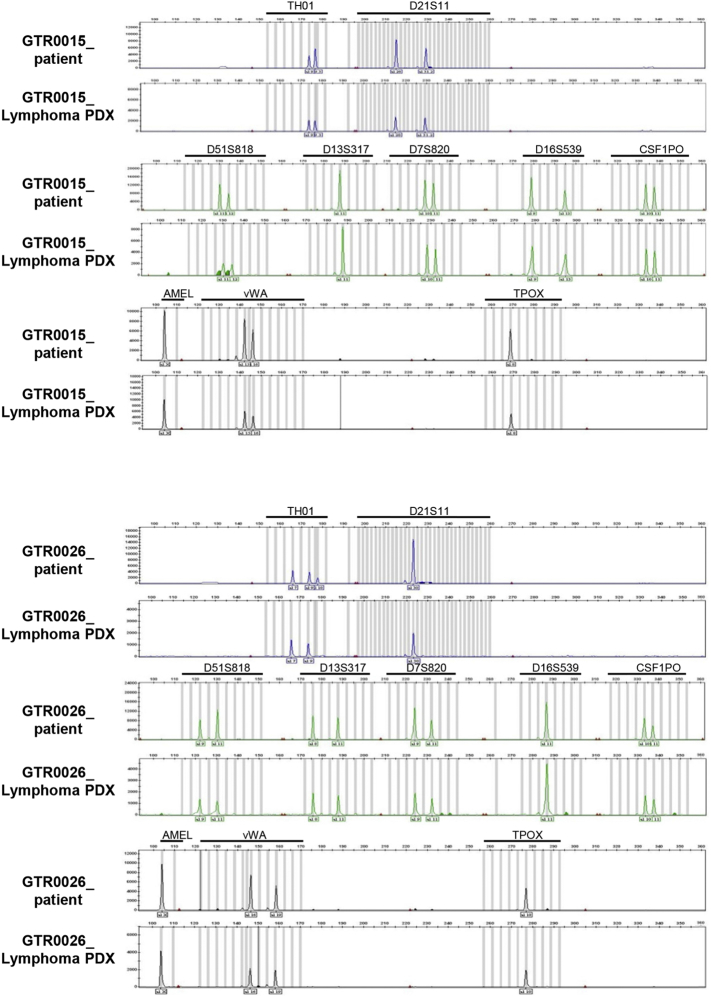
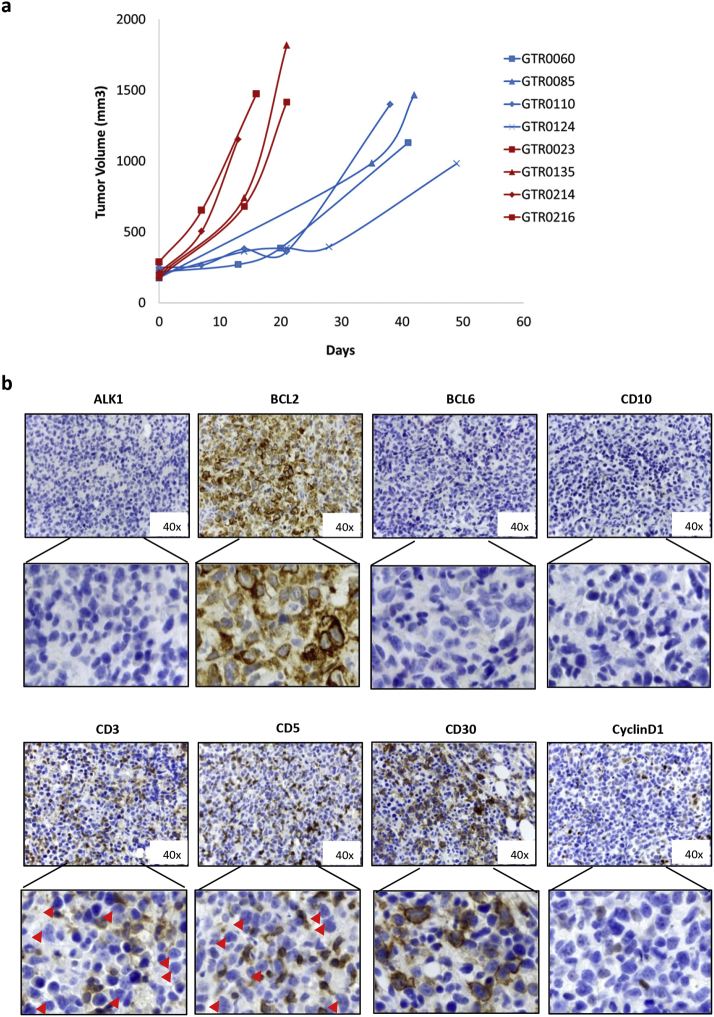
As it is known that NOD SCID mice can develop murine lymphomas at high frequency, we evaluated the species of origin of the neoplastic cells. Short tandem repeats (STR) analysis of the PDX and of the respective original gastric tumor showed that they shared the same STRs (representative examples are shown in Supplementary Figure 1), proving that the lymphomas were patient-derived. Moreover, as displayed in Figure 1D, these tumors were reactive with antibodies specifically recognizing the human CD20 antigen (18/18 analyzed samples), thus confirming that they derived from human B-lymphocytes. As expected, all the lymphomas were negative for a pan-type cytokeratin staining and showed positivity for CD79, confirming the B-cell derivation of tumor cells (Figure 1D). Moreover, lymphomas displayed a high MIB-1 (KI-67) index, indicative of a high proliferation rate (Figure 1D). This is consistent with the observed growth kinetic; indeed, lymphoma PDXs grew significantly faster than PDXs maintaining the carcinoma histology, as the average tumor growth rate in lymphoma PDXs was 70.7 mm3/day, while in Gastric PDXs it was 24.8 mm3/day (P = .00016; Supplementary Figure 2A).
Supplementary Figure 1.
Lymphoma PDXs are of human origin.
Electropherogram of two representative Lymphoma PDXs compared to their respective gastric cancer of origin. Ten STR loci were analyzed. As shown, the paired STRs demonstrate identical alleles, confirming that the Lymphomas derived from the patients' gastric cancers.
Supplementary Figure 2.
Growth rate and immunophenotyic analysis of lymphoma PDXs.
a. Growth curves of 4 representative lymphoma PDXs (red lines) and 4 representative Gastric PDXs (blue lines). As shown, Lymphoma PDXs grew significantly faster than Gastric PDXs (average tumor growth rate in Lymphoma PDXs was 70.7 mm3/day; average tumor growth rate in Gastric PDXs was 24.8 mm3/day; P = .00016 (t-test)). b. lmmunohistochemical analysis of one representative Lymphoma PDX for ALK1, BCL2, BCL6, CD10, CD3, CD5, CD30 and Cyclin D1. Lowers panels show enlargement of areas of the upper panels. Red arrows indicate large lymphoma cells, which appear negative for the staining.
A deeper immunophenotyping of the samples revealed that the developed lymphomas contained CD3/CD5 negative large B lymphoma cells and a minority of CD3/CD5 positive small reactive T Lymphocytes, as frequently found in human B cell lymphomas.19 Moreover, lymphoma cells were CD30 and Bcl2 positive, CD10, ALK1, CyclinD1 and Bcl-6 negative (Supplementary Figure 2B).
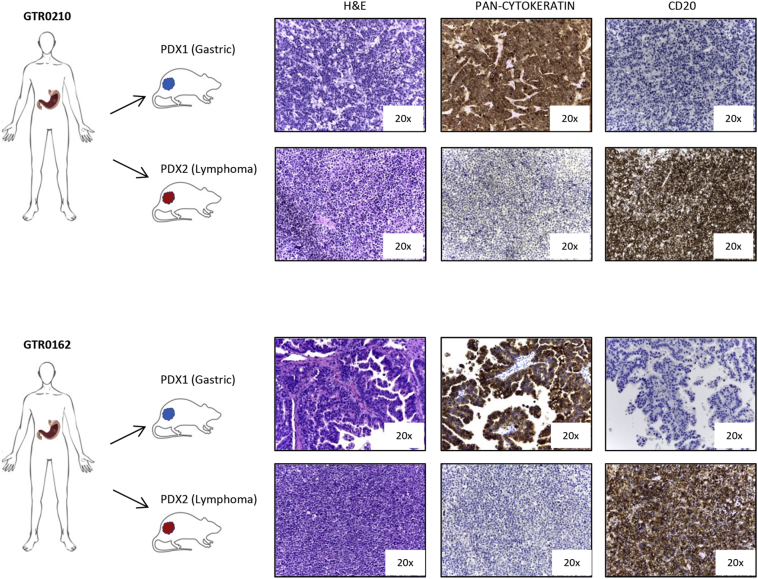
During the generation of our PDX platform, usually the patient-derived gastric carcinoma samples have been implanted in parallel into two mice. In nearly all cases, first generation PDXs developed either carcinomas or lymphomas; however, in 5 cases the same sample generated one gastric carcinoma (counted in the Gastric PDX group) and one lymphoma (counted in the Lymphoma PDX group) (Supplementary Figure 3).
Supplementary Figure 3.
Gastric cancer samples implanted in two mice can separately give rise to a lymphoma and a gastric PDXs.
The figure shows 2 cases (GTR0210 and GTR0162) in which one of the implanted mice developed a gastric carcinoma and the other one developed a lymphoma. H&E, pan-cytokeratin and CD20 staining confirmed the different histology of the PDXs originated from the same gastric tumor.
Lymphoma Onset Does Not Correlate With the Level of Lymphocyte Infiltration, the Histotype of the Original Gastric Tumor, or With Patient Outcome
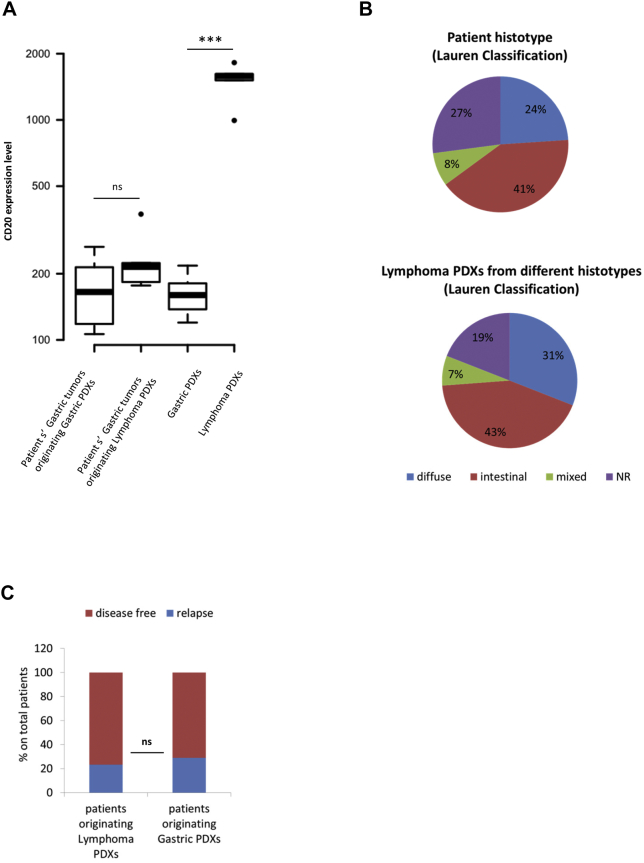
As PDX lymphomas originate from human lymphocytes, we hypothesized the existence of a relationship between the number of lymphocytes (TILs) infiltrating the original tumor and lymphoma development in mice. We thus performed transcriptomic analysis of the surgically removed gastric tumors and analyzed CD20 expression. As shown in Figure 2A, no correlation was observed between the amount of B lymphocytes present in the stroma of the gastric carcinoma and lymphoma onset, suggesting that the amount of immune cells infiltrating the tumor is not directly correlated with the probability of developing lymphomas.
Figure 2.
Lymphoma onset does not correlate with the degree of lymphocyte infiltration or with the histotype of the original gastric tumor or with patient outcome. A. The Boxplot reports the distribution of human CD20 expression level, extrapolated from gene array analysis, in primary patients’ gastric tumors originating either Gastric or Lymphoma PDXs, and in the corresponding Gastric or Lymphoma PDXs. CD20 expression level (directly correlated with the amount of tumor infiltrating lymphocytes) was not significantly different in patients’ gastric tumors originating either Gastric or Lymphoma PDXs (P = .1179, Wilcoxon test). As expected, gastric PDXs showed few human tumor infiltrating lymphocytes, while lymphoma PDXs displayed the highest CD20 expression level (P = .0002, Wilcoxon test). B. Upper graph: histotypes of primary patients’ gastric carcinomas implanted in mice (according to Lauren classification); lower graph: histotypes of patients’ gastric carcinomas originating Lymphoma PDXs. As shown, lymphoma PDXs originated from the different histotypes. NR= Lauren Classification not reported. No significant difference in lymphoma onset between diffuse and intestinal histotypes was found (P = 0.5457, Fisher's exact test). C. The graph represents the follow up (updated at January 2017) of donor patients undergoing surgery between 2013 and 2015. Among the patients whose tumor originated a Lymphoma PDX, 8 out of 33 (for whom follow up was available) relapsed (24%). Among patients whose tumor originated a Gastric PDX, relapse was observed in 15 out of 45 patients (33%). The difference was not statistical significant (P = .7828; Fisher’s exact Test).
Similarly, no significant correlation was found between the histotype of the original gastric tumor and the onset of PDX lymphomas; indeed, as shown in Figure 2B, lymphomas originated from all the Lauren’s histotypes (intestinal, diffuse or mixed tumors), at proportional frequency. Finally, no correlation was found with patient outcome. Follow up of patients undergoing surgery between 2013 and 2015 revealed that their relapse rate was independent from the type of originated PDX (Figure 2C).
Genomic and Transcriptomic Analysis of PDX Lymphomas
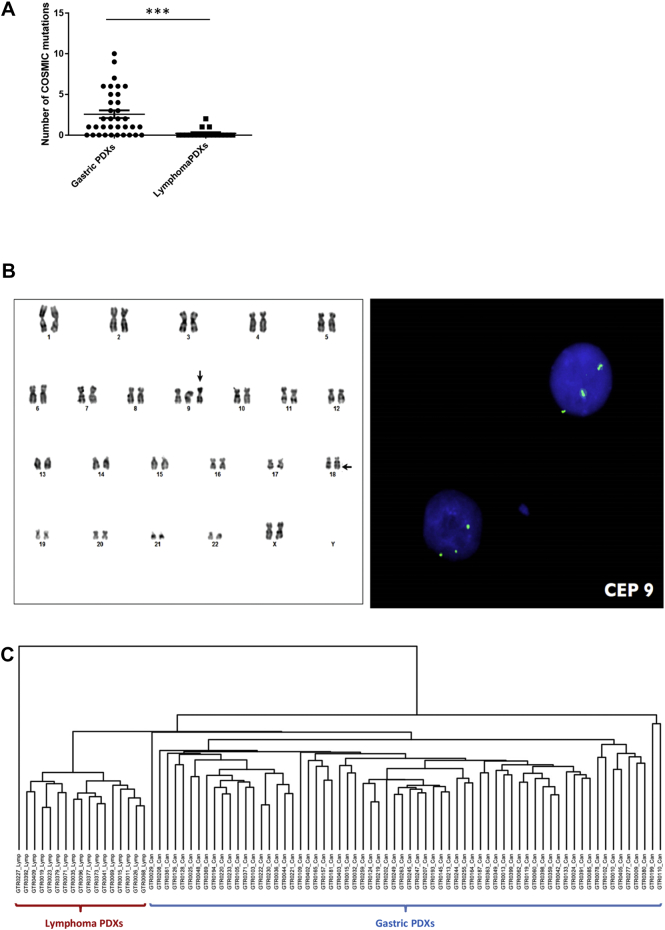
To further characterize lymphoma PDXs we performed genetic and transcriptomic analyses. Genomic sequencing of a panel of 250 genes revealed the presence of a significant number of genomic alterations, although at a significantly lower level compared to gastric carcinoma PDXs (Figure 3A). Interestingly, all the analyzed lymphomas (20/20) harbored at least one loss-of-function mutation in ARID1A/B or in ARID2 genes, components of the SWI-SNF chromatin remodeling complex, which takes part in somatic cells reprogramming.[20], [21] Interestingly, ARID1A has been already reported as a recurrently mutated gene in human Follicular Lymphoma22 and in Diffuse large B-cell lymphoma.23
Figure 3.
Genomic and transcriptomic analysis of PDX lymphomas. A. Targeted sequencing of a panel of 250 genes was performed on 35 gastric PDXs and 18 lymphoma PDXs. The graph represents the number of COSMIC mutations identified in each PDX. Mutational burden was significantly higher in gastric PDXs compared to lymphomas (****P < .0009, Unpaired t-test). B. Karyotype of cells derived from a lymphoma PDX. Arrows indicate the trisomy of chromosome 9 (vertical arrow) and the t(X;18)(q25;q21.3) translocation (horizontal arrow). C. Unsupervised hierarchical clustering performed on transcriptomic analysis of 66 gastric PDXs and 17 lymphoma PDXs. The analysis revealed the strong partition of the two PDX subgroups.
Cytogenetic analysis of lymphoma cells evinced the presence of chromosomal aberrations, such as the trisomy of chromosome 9 and the t(X;18)(q25;q21.3) translocation shown in Figure 3B. FISH analysis revealed the hemizygous loss of BCL2 gene (18q21.3) as a consequence of the t(X;18) translocation.
Unsupervised hierarchical clustering performed on transcriptomic analysis of both lymphoma and carcinoma PDXs completely separated lymphoma propagation from gastric cancer propagation, further proving their different cellular origin (Figure 3C).
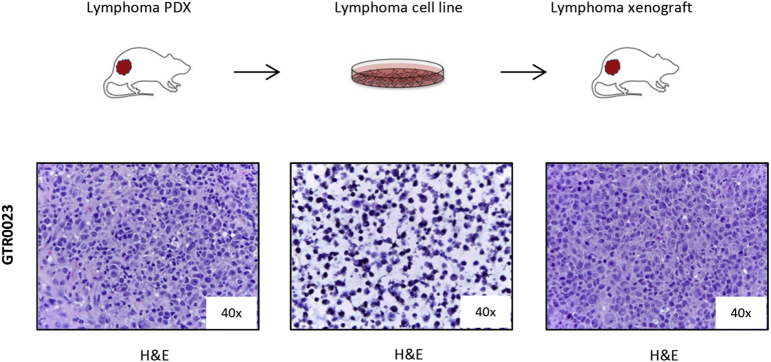
Finally, several cell lines were derived in culture from lymphoma PDXs; these cells were able to indefinitely grow in vitro and to originate tumors when re-injected in mice (Supplementary Figure 4).
Supplementary Figure 4.
Cell lines derived in culture from lymphoma PDXs are tumorigenic.
The figure shows the H&E staining of one representative Lymphoma POX (FFPE, left panel), the corresponding lymphoma cell line derived in culture {cyto-spin, middle panel) and the lymphoma xenograft originated upon s.c. injection of the lymphoma cell line in mice (FFPE, right panel).
Lymphoma PDXs are EBV Positive and Display Monoclonal Immunoglobulin Heavy Chain (IgH) Rearrangements
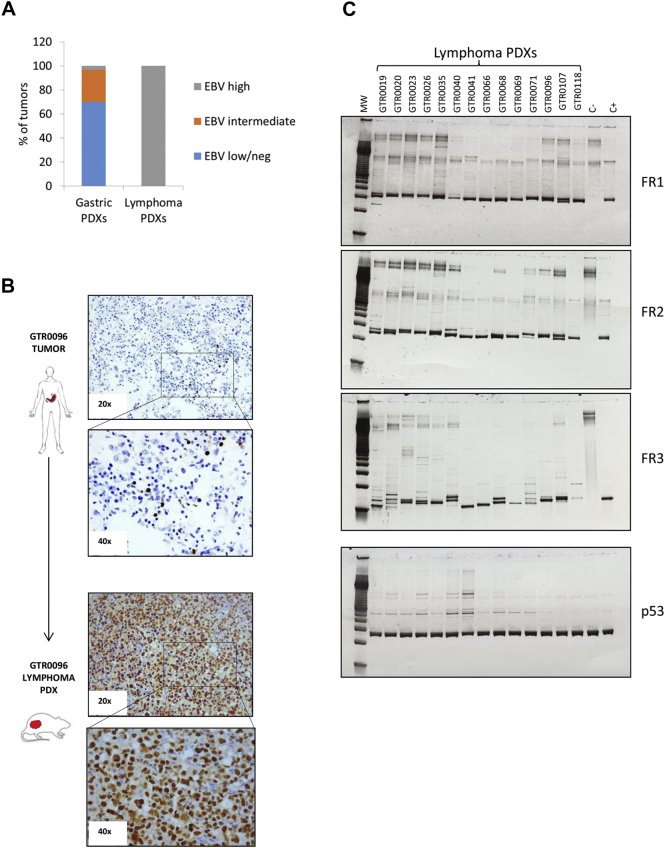
As lymphomagenesis in immune-depressed patients is known to be related with EBV positivity, we assessed the EBV status of lymphoma PDXs. All the analyzed lymphomas turned out strongly EBV positive (Figure 4A); however, no correlation was observed between EBV positivity of primary tumors and lymphoma onset. Indeed, two primary tumors that were intensely positive for the virus originated gastric PDXs and not lymphomas (data not shown); conversely, most lymphomas derived from gastric tumors that were EBV negative, according to routine Real Time scoring procedures (Supplementary Table 2). In order to understand if EBV negative scoring of these tumors was due to the paucity of EBV+ cells within the gastric carcinoma tissue, we performed in-situ hybridization (ISH) for the Epstein Barr Virus encoded RNA (EBER), which is abundantly expressed in latent EBV infection.24 As shown in Figure 4B, rare EBV+ cells could be detected in some gastric carcinomas that originated lymphoma in immunocompromised mice. However, no EBV+ cells could be detected in the slices of other gastric carcinomas generating lymphoma (data not shown). We hypothesize that in EBV cases scored negative by ISH, rare positive cells could be present in other parts of the tumor, suggesting that the analysis of the whole carcinoma should be performed to definitively assess the presence of EBV+ lymphocytes. When the presence of EBV DNA was investigated (by real-time-based assays) in white blood cells obtained from gastric cancer patients, no significant difference in viral copies was observed between those which originated gastric or lymphoma PDXs (not shown). Altogether, these results indicate that the EBV status of cells present in the original tumor, cannot be routinely exploited to predict lymphoma occurrence in mice.
Figure 4.
Lymphoma PDXs are all EBV positive and display monoclonal Ig Heavy Chain rearrangements. A. EBV status was assessed in Gastric and Lymphoma PDXs through Real-Time quantification on genomic DNA, according to the protocol adopted by the Laboratory Medicine of the Hospital. PDXs were classified as EBV high (with high EBV burden, >1000, Equivalent EBV Genomes/100000 cells (gEq × 10e4 cells), EBV intermediate (75-1000 gEq × 10e4 cells) or EBV low/neg (<75 gEq × 10e4 cells). All the analyzed Lymphoma PDXs (n = 24) displayed high EBV burden, compared to 3% of the Gastric PDXs (n = 30). B. Representative in-situ hybridization (ISH) for the Epstein Barr Virus encoded RNA (EBER) in a patient’s gastric carcinoma (upper part) and in the corresponding lymphoma PDX (lower part). While only rare EBV+ cells were detected in the patient’s sample, strong positivity was observed in the lymphoma. C. Clonality of the B cell lymphoproliferative processes was evaluated in 14 representative Lymphoma PDXs. PCR amplification of rearranged VDJ segments (FR1-JH, FR2-JH and FR3-JH) in the hypervariable region of the immunoglobulin heavy chain, using multiple primers that hybridize with conserved regions in the genes, was performed on Lymphoma PDX gDNAs. The DNA quality was checked for all samples using the p53 control gene. C-: PCR performed on genomic DNA of a patient without monoclonal Ig heavy chain rearrangement (negative control); C+: PCR performed on genomic DNA of a patient with monoclonal Ig heavy chain rearrangement (positive control). As shown, all the Lymphoma PDXs displayed monoclonal Ig heavy chain rearrangements in at least 1 PCR reaction.
To evaluate the clonal origin of these lymphatic neoplasms we looked for IgH rearrangements in the neoplastic cells. As shown in Figure 4B, most of the lymphomas displayed a clonal IgH rearrangement, proving their monoclonal origin. However, no IgH monoclonal rearrangement could be detected in the corresponding primary gastric tumors or in patient’s blood (data not shown).
Lymphomas Originate at Lower Frequency From Metastatic Than Primary Tumors
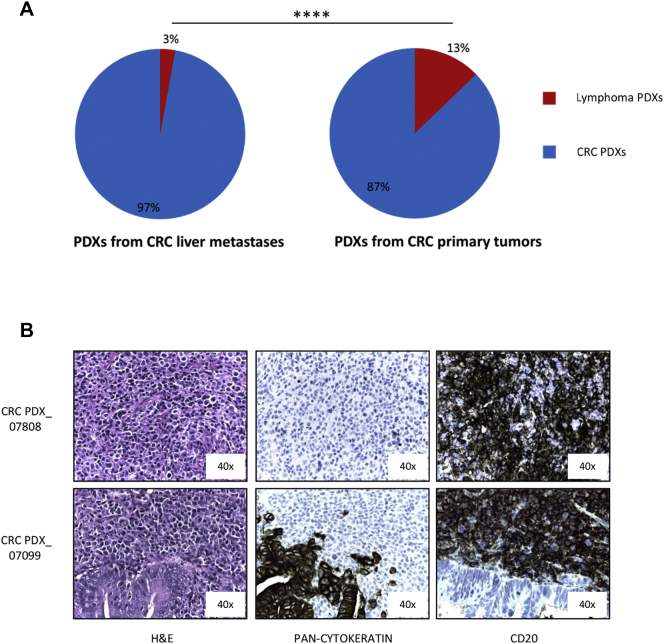
As the gastrointestinal mucosa is rich in immune cells (and among them of B lymphocytes), we wondered if lymphomagenesis in PDXs might be influenced by the tissue from which the tumor is surgically removed. Since metastatic gastric PDXs were not present in our platform, we took advantage from colon cancer PDXs available in our Institute, derived from both primary and liver metastatic tumors.25 While only 3% of grafted metastatic colon tumors generated human lymphomas (7/246), this happened only in around 13% of primary colon cancer PDXs (25/195, P < .0001, Fisher's exact test) (Figure 5A). Also in these cases all the tested lymphomas were CD20+ (Figure 5B) and strongly EBV+ (>1000 Equivalent EBV Genomes/100000 cells, data not shown).
Figure 5.
Lymphoma PDXs arise at higher frequency from primary tumors compared to metastases. A. Percentage of Lymphoma PDXs onset in a PDX platform obtained from NOD/SCID mice implant of liver metastases from colorectal cancer (7/246, left graph) or of primary colorectal tumors (25/195, right graph). (P < .0001, Fisher's exact test). B. Immunohistochemical analysis of two representative lymphomas (one lymphoma –upper figures- and one Mixed PDX –lower figures-) derived from primary colorectal cancers. H&E, pan-cytokeratin and CD20 staining are shown.
Lymphomagenesis in PDXs is Sensitive to Rituximab Treatment
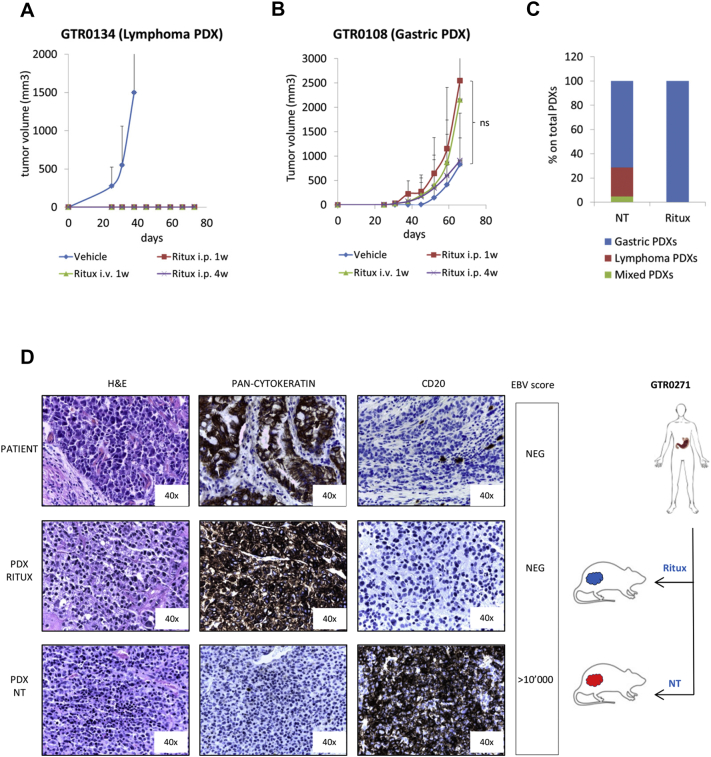
Preclinical and clinical experience has shown that human lymphomas are sensitive to treatment with rituximab, an anti-CD20 monoclonal antibody.[26], [27] We thus verified if rituximab treatment could impair lymphoma growth in mice. As shown in Figure 6A, when the drug was administered intra-peritoneum for 4 weeks, starting 4 days after implantation of Lymphoma PDXs, it completely impeded lymphoma growth. The same result was obtained with a 1-week treatment (intra-peritoneum or intra-vein administration). On the other hand, rituximab treatment did not hamper the growth of gastric carcinomas (Figure 6B). Interestingly, in a case where the engrafted tumor gave rise to a Mixed PDX (carcinoma and lymphoma in different areas of the tumor mass (Supplementary Figure 5, middle panels), rituximab treatment after re-implantation allowed growth of a tumor with features of pure adenocarcinoma (Supplementary Figure 5, lower panels). Rituximab treatment thus succeeded in rescuing PDX models from EBV positive contaminant B-cells.
Figure 6.
Rituximab treatment prevents lymphoma onset without affecting Gastric PDX engraftment. A. A Lymphoma PDX (GTR0134) was subcutaneously implanted in 12 NOD/SCID mice. Four days after implant, 3 animals were treated with physiologic solution (‘Vehicle’), 3 with Rituximab i.p. 25 mg/kg, 3 times/week for 1 week (‘Ritux i.p.1w’), 3 with Rituximab i.p. 25 mg/kg, 3 times/week for 1 week and once a week for the following 3 weeks (Ritux i.p.4w), 3 with Rituximab i.v. 10 mg/kg, 3 times/week for 1 week (Ritux i.v.1w). The graph shows the tumor growth curves from the day of implant (day 0). Rituximab treatment completely inhibited lymphoma growth in mice. B. A gastric PDX (GTR0108) was subcutaneously implanted in 12 NOD/SCID mice. Four days after the implant, mice were divided in 4 groups and treated as in A. The graph shows the tumor growth curves from the day of implant (day 0). Rituximab treatment did not hamper gastric carcinoma growth. C. Gastric tumors received from March to August 2016 were implanted in at least 2 NOD/SCID mice; one of them was untreated (‘NT’), one was treated with Rituximab i.p. 25 mg/kg 3 times/week for 1 week and once a week for the following 3 weeks (Ritux). The graph reports the percentages of Gastric, Lymphoma and Mixed PDX onset in mice undergoing the different treatments. As shown, Rituximab treatment completely prevented the onset of lymphomas (18/18 Gastric PDXs in the Rituximab group). D. The gastric tumor from patient GTR0271 (upper panels) originated a Gastric PDX when grown in a Rituximab-treated mouse (middle panels) and a Lymphoma PDX when grown in an untreated animal (lower panels). H&E, pan-cytokeratin and CD20 staining confirmed the different histology of the PDXs. The EBV score, negative in the donor tumor and in the Gastric PDX and highly positive in the Lymphoma PDX, is also reported in the figure.
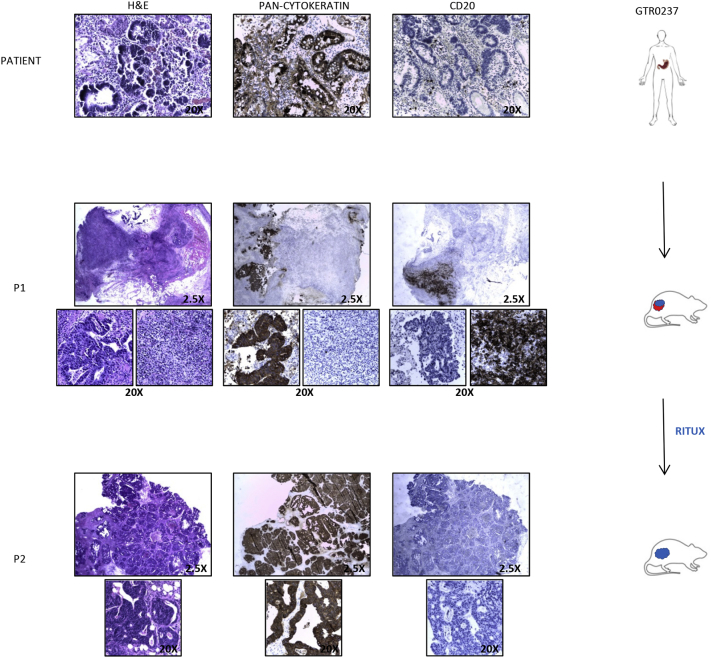
Supplementary Figure 5.
Rituximab treatment after re-implantation of a Mixed PDXs prevents lymphoma formation.
The gastric tumor from patient GTR0237 (upper panels) originated a Mixed Lymphoma in the first passage in mouse (P1, middle panels and enlargements). The mixed tumor was s.c transplanted in a second mouse (P2, lower panels and enlargements) treated with Rituximab i.p. 25 mg/kg 3 times/week for 1 week and once a week for the following 3 weeks (Ritux). As shown, Rituximab treatment allowed growth of a tumor with features of pure adenocarcinoma. H&E, pan-cytokeratin and CD20 staining confirmed the different histology of the P1 and P2 PDXs.
We also wondered if rituximab might be used in a preventive setting, to avoid the onset of lymphomas, thus increasing the percentage of carcinoma engraftment. From March to August 2016 each received tumor was therefore implanted in at least 2 mice, one of them treated for 4 weeks with rituximab, starting at day 4 after implantation. Among the rituximab-treated mice, none developed lymphomas (Figure 6C), while the rate of gastric PDX engraftment was not significantly different between treated and untreated mice (data not shown). Interestingly, the same primary gastric tumor could originate a lymphoma when grafted in the untreated mouse and a gastric carcinoma when grown in the rituximab-treated mouse (Figure 6D). This demonstrates that the gastric cancer samples originating lymphoma PDXs, in absence of rituximab, are not devoid of engraftment potential and suggests that rituximab treatment can be used to prevent lymphomagenesis and increase carcinoma engraftment rate.
Discussion
Differently from other tumors, for which many therapeutic options are currently available, only two target therapies have been approved for gastric cancer so far.[3], [4], [5] Over the past years, many molecular drugs entered clinical trials for this kind of cancer but, with the exception of Trastuzumab and Ramucirumab, they all failed, suggesting the need for a more accurate patient selection and for more solid preclinical bases.6
The use of PDXs as an emerging translational model for cancer research has raised interest in the last years,28 and could help improve gastric cancer therapy. Indeed, PDX models usually retain the histologic and genetic features of the tumor of origin and have been shown to be predictive of clinical outcome.1 Therefore, they are widely used to perform preclinical trials evaluating drug response and to identify biomarkers and novel therapeutic targets. PDX generation requires the close interaction between surgeons, pathologists and researchers, as tumor engraftment demands accurate manipulation of the samples and very strict timing. Nevertheless, the ability of the tumor to engraft varies among tumor types.28 Different non-immunocompetent strains of mice can be used for this procedure, all of them offering pro and cons. The most immune-depressed ones usually display a higher graft rate, but also a higher propensity to develop lymphoproliferative diseases. Formerly, lymphomas of mouse origin were described to take place either in the site of engraftment or in different organs.29 More recently, in depth analysis of lymphatic neoplasms generated in mice engrafted with human carcinoma samples showed that many of them were of human origin.[8], [30], [31], [32] The onset of tumors of a histotype different from the original one represents a risk for the correct interpretation of ‘xenotrial’ results. Indeed, if researchers are not aware of this issue, the potential development of lymphomas, instead of the awaited carcinomas, could impact on the results of the preclinical trials evaluating drug activity. In fact, as expected, the human-derived lymphoproliferative diseases developed in PDXs do not share with the tumor of origin the molecular alterations that justify the administration of a certain targeted therapy. Thus, this could result in the failure of the ‘xenotrial’, not due to the lack of activity of the drug, but rather to inappropriate PDX inclusion.
In our work we evaluated a platform of gastric PDXs in NOD SCID mice and found that around 34% of engrafted tumors were lymphomas of human origin. These tumors have been diagnosed by the pathologists as large B cell lymphomas and were all strongly positive for the EBV virus. However, no correlation could be identified between EBV positivity of the original tumor (or of patients’ white blood cells) and the onset of lymphomas in mice. Most EBV+ lymphomas derived from EBV negative gastric carcinomas. Considering the monoclonal origin of the lymphomas, it is possible that in some tumors, scored as EBV negative by real-time PCR, the number of EBV+ lymphocytes was below the detection threshold; however, these rare genetically/genomically altered lymphocytes might have had a selective advantage in an immune-depressed environment. This hypothesis is supported by the identification, through ISH analysis, of rare EBV+ cells in some gastric tumor samples originating lymphomas in immunocompromised mice. We cannot exclude that, in the cases where ISH has not identified EBV+ cells, few positive lymphocytes might be present in other parts of the tumor and in particular in the small tumor sample implanted in the mouse. Finally, in two cases, EBV+ gastric carcinomas originated gastric PDXs and not lymphomas; in these cases it is likely that gastric epithelial cells (and not lymphocytes) presented latent EBV infection. Altogether, these results indicate that the EBV status of cells present in the original tumor cannot be routinely exploited to predict lymphoma occurrence in mice.
We did not find any correlation between lymphoma onset in mice and clinical outcome of the original patients. Since none of them developed a lymphoproliferative disease, it is likely that a deep immunodepressive condition, rarely found in patients, is mandatory to unleash the oncogenic potential of the altered B cells. It has also to be noted that the life expectancy of advanced gastric cancer patients, unfortunately, is not often long enough to allow for lymphoma appearance.
From a molecular and biological point of view we found that lymphomas: (i) were of monoclonal origin as they presented monoclonal rearrangements of immunoglobulin heavy chains; (ii) in some cases displayed evident chromosomal abnormalities; (iii) were able to originate tumorigenic cell cultures, maintaining the same molecular features; (iv) displayed a lower mutational tumor burden compared to gastric adenocarcinomas; (v) always presented mutations of ARID1/2 genes, frequently identified in follicular and diffuse large B cells lymphomas; (vi) showed a distinct expression profile compared to gastric adenocarcinomas; (vii) grew faster than gastric adenocarcinomas. The presence of monoclonal Ig rearrangements is not an absolute sign of neoplastic transformation as it can be found in pathologies such as MGUS (Monoclonal Gammopathies of Undetermined Significance). However, the concomitant identification of monoclonal Ig rearrangements, chromosomal rearrangements and several DNA point mutations, joined with the ability of lymphoma PDXs to be serially transplanted, strongly support the neoplastic origin of these processes.
As B lymphocytes are the cells of origin of these lymphomas, we expected to find a connection between the amount of B cells infiltrating the implanted tumor and lymphomas onset. However, this was not the case as the evaluation of CD20 expressing cells in the primary tumor did not correlate with the ability to originate lymphomas.
Nevertheless, while the gastro-intestinal mucosa is known to be rich in immune cells, this is not true for metastatic sites as liver. Since in our platform we did not have the opportunity to generate PDXs from metastatic gastric tumors, we took advantage from a colorectal PDX platform generated in our institute.25 Interestingly, we found that while primary colo-rectal tumors originated EBV positive lymphomas at a relatively high frequency (13%), very few lymphoproliferative disorders of human origin were originated from implanted metastatic tumors. Since immune cells are much less represented in the liver parenchyma compared with intestinal mucosa, these experiments suggest that, above a certain threshold, the immune component of the tumor tissue of origin might contribute to the lymphogenic potential in mice. In other words, the number of latent EBV-infected B cells, affecting lymphoma PDX formation, could vary significantly in tumors of different anatomic origin, such as stomach, colon, and liver. This novel observation further reinforces the idea that the microenvironment of tumor cells strongly conditions the onset of lymphomas.
Finally, as the development of lymphomas rather than carcinomas results in loss of valuable PDX models, we aimed at preventing their onset. Rituximab is a monoclonal antibody directed against CD20, currently used in the clinic to treat B cell lymphomas.[26], [27] As it does not react with mouse B lymphocytes, we tested its therapeutic and preventive ability in our PDXs. Indeed, we show that a short rituximab treatment, started just after mice transplantation, was able to prevent lymphoma onset in PDXs without affecting growth of adenocarcinomas. Moreover, rituximab treatment of mixed (carcinoma and lymphoma) samples after re-implantation allowed the growth of tumors with features of pure adenocarcinoma, providing an important strategy to successfully rescue PDX models from EBV positive contaminant B-cells. Choi and colleagues reported that, using nude mice (lacking T-cell function only), they could prevent lymphoma formation during establishment of gastric cancer PDX models.33 However, changing the mouse strain in already established PDX platforms is not always possible or recommended, since it is well known that microenvironment can influence tumor functions.34 As some works reported that human lymphomas originate from tumor grafts of several histotypes (although at different frequency),[30], [31], [32] we propose rituximab treatments as an efficient and relatively easy procedure to prevent lymphoma development and to increase adenocarcinoma engraftment rate in PDX models.
Conclusions
In conclusion, in a wide gastric cancer PDX platform, we show that 30% of immunocompromised mice implanted with gastric tumors develop human-derived monoclonal lymphomas instead of gastric cancer, a percentage much higher than the one observed in PDXs derived from different tumor histotypes. Moreover, we demonstrate that preventive rituximab treatment impairs lymphomas onset, without affecting gastric tumor uptake. This is critical to avoid the execution of preclinical trials on inappropriate material.
The following are the supplementary data related to this article.
The Table shows the panel of immunohistochemical markers analyzed and of the antibodies employed.
EBV score (Equivalent EBV Genomes/100000 cells) in primary gastric carcinomas and in the corresponding Lymphoma PDXs originated in NOD/SCID mice.
Supplementary Materials and Methods
Acknowledgments
Acknowledgements
We thank Dr. L.Trusolino and all our colleagues for helpful scientific discussion; Drs Porporato, Buscarino and Montone for technical support with gene array and Cell-ID; animal facility employees; Francesco Fesi for EBV analysis; Dr. Natale for critical manuscript reading. SG, SC, EM and ABertotti are EuroPDX Consortium members. All the GIRCG (Gruppo Italiano Ricerca Cancro Gastrico) members. This work was funded by the Italian Association for Cancer Research (AIRC); IG grant 15464 to SG and Fondazione Piemontese per la Ricerca sul Cancro (ONLUS) 5 X 1000 Fondi Ministero della Salute 2013 to A. Sottile. Genomic sequencing was supported by the Schottenstein Fund for Gastric Cancer Research to ABass.
Author contributions
SG and SC conceived and supervised the study, contributed to design the experiments, wrote the manuscript. MD, UF, GS, SMolfino, GDM, RR, MDS, DM, LS, SR, GP, FR provided the patient material, MC, SD, SMenegon, MA, CM, TC, ABertotti, FS performed experiments; ASapino, PC, IS, AP performed the pathologic analysis; LC performed FISH analysis; ASottile and MM performed EBV evaluation and analysis of Ig Heavy Chain rearrangements; CI, EM and ABass performed transcriptomic and genomic analysis. All the authors revised the manuscript.
Disclosures
All institutional and national guidelines for the care and use of laboratory animals were followed.
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Conflict of interest: No conflict of interest.
Array data available in: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=snspycqynfyhful&acc=GSE98708.
Contributor Information
Simona Corso, Email: simona.corso@unito.it.
Silvia Giordano, Email: silvia.giordano@unito.it.
References
- 1.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Mælandsmo GM. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C. A molecularly annotated platform of patient-derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 3.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, Safran H, dos Santos LV, Aprile G, Ferry DR. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–39. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 5.Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, Hironaka S, Sugimoto N, Lipatov O, Kim TY. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–1235. doi: 10.1016/S1470-2045(14)70420-6. [DOI] [PubMed] [Google Scholar]
- 6.Corso S, Giordano S. How can gastric cancer molecular profiling guide future therapies? Trends Mol Med. 2016;22:534–544. doi: 10.1016/j.molmed.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Fujii E, Kato A, Chen YJ, Matsubara K, Ohnishi Y, Suzuki M. Characterization of EBV-related lymphoproliferative lesions arising in donor lymphocytes of transplanted human tumor tissues in the NOG mouse. Exp Anim. 2014;63:289–296. doi: 10.1538/expanim.63.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K, Ahmed S, Adeyi O, Dick JE, Ghanekar A. Human solid tumor xenografts in immunodeficient mice are vulnerable to lymphomagenesis associated with Epstein-Barr virus. PLoS One. 2012;7:e39294. doi: 10.1371/journal.pone.0039294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JI, Kimura H, Nakamura S, Ko YH, Jaffe ES. Epstein-Barr virus-associated lymphoproliferative disease in non-immunocompromised hosts: a status report and summary of an international meeting, 8-9 September 2008. Ann Oncol. 2009;20:1472–1482. doi: 10.1093/annonc/mdp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbone A, Gloghini A, Dotti G. EBV-associated lymphoproliferative disorders: classification and treatment. Oncologist. 2008;13:577–585. doi: 10.1634/theoncologist.2008-0036. [DOI] [PubMed] [Google Scholar]
- 11.Galimi F, Torti D, Sassi F, Isella C, Corà D, Gastaldi S, Ribero D, Muratore A, Massucco P, Siatis D. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: response to met inhibition in patient xenografts and pathologic correlations. Clin Cancer Res. 2011;17:3146–3156. doi: 10.1158/1078-0432.CCR-10-3377. [DOI] [PubMed] [Google Scholar]
- 12.Shimodaira RSaH. pvclust: Hierarchical clustering with P-values via multiscale bootstrap resampling R package version 2.0-0. 2015. https://CRAN.R-project.org/package=pvclust
- 13.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Team RC. 2014. R: A Language and environment for statistical computing: R foundation for statistical computing. [Google Scholar]
- 15.Marty GD. Blank-field correction for achieving a uniform white background in brightfield digital photomicrographs. Biotechniques. 2007;42:716. doi: 10.2144/000112488. [8, 20] [DOI] [PubMed] [Google Scholar]
- 16.Apicella M., Migliore C, Capelôa T, Menegon S, Cargnelutti M, Degiuli M, Sapino A, Sottile A, Sarotto I, Casorzo L. Dual MET/EGFR therapy leads to complete response and resistance prevention in a MET-amplified gastroesophageal xenopatient cohort. Oncogene. 2017;36:1200–1210. doi: 10.1038/onc.2016.283. [DOI] [PubMed] [Google Scholar]
- 17.McGowan-Jordan J, Simons A, Schmid M. Karger Publisher; 2016. ISCN 2016 An International System for Human Cytogenomic Nomenclature. [Google Scholar]
- 18.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng CS, Chan JK, Hui PK, Lau WH. Large B-cell lymphomas with a high content of reactive T cells. Hum Pathol. 1989;20:1145–1154. doi: 10.1016/s0046-8177(89)80004-8. [DOI] [PubMed] [Google Scholar]
- 20.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 21.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, Fox H, Jacobi K, Saiya-Cork K, Bixby D. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, Dunphy C, Choi W, Au WY, Srivastava G. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambinder RF, Mann RB. Epstein-Barr-encoded RNA in situ hybridization: diagnostic applications. Hum Pathol. 1994;25:602–605. doi: 10.1016/0046-8177(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 25.Zanella ER, Galimi F, Sassi F, Migliardi G, Cottino F, Leto SM, Lupo B, Erriquez J, Isella C, Comoglio PM. IGF2 is an actionable target that identifies a distinct subpopulation of colorectal cancer patients with marginal response to anti-EGFR therapies. Sci Transl Med. 2015;7:272ra12. doi: 10.1126/scitranslmed.3010445. [DOI] [PubMed] [Google Scholar]
- 26.Maloney DG, Grillo-López AJ, White CA, Bodkin D, Schilder RJ, Neidhart JA, Janakiraman N, Foon KA, Liles TM, Dallaire BK. IDEC-C2B8 (Rituximab) anti-CD20 monoclonal antibody therapy in patients with relapsed low-grade non-Hodgkin's lymphoma. Blood. 1997;90:2188–2195. [PubMed] [Google Scholar]
- 27.Oldham RK, Dillman RO. Monoclonal antibodies in cancer therapy: 25 years of progress. J Clin Oncol. 2008;26:1774–1777. doi: 10.1200/JCO.2007.15.7438. [DOI] [PubMed] [Google Scholar]
- 28.Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, Bruna A, Budinská E, Caldas C, Chang DK. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17:254–268. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 29.Prochazka M, Gaskins HR, Shultz LD, Leiter EH. The nonobese diabetic scid mouse: model for spontaneous thymomagenesis associated with immunodeficiency. Proc Natl Acad Sci U S A. 1992;89:3290–3294. doi: 10.1073/pnas.89.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John T, Yanagawa N, Kohler D, Craddock KJ, Bandarchi-Chamkhaleh B, Pintilie M, Sykes J, To C, Li M, Panchal D. Characterization of lymphomas developing in immunodeficient mice implanted with primary human non-small cell lung cancer. J Thorac Oncol. 2012;7:1101–1108. doi: 10.1097/JTO.0b013e3182519d4d. [DOI] [PubMed] [Google Scholar]
- 31.Bondarenko G, Ugolkov A, Rohan S, Kulesza P, Dubrovskyi O, Gursel D, Mathews J, O'Halloran TV, Wei JJ, Mazar AP. Patient-Derived Tumor Xenografts Are Susceptible to Formation of Human Lymphocytic Tumors. Neoplasia. 2015;17:735–741. doi: 10.1016/j.neo.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wetterauer C, Vlajnic T, Schüler J, Gsponer JR, Thalmann GN, Cecchini M, Schneider J, Zellweger T, Pueschel H, Bachmann A. Early development of human lymphomas in a prostate cancer xenograft program using triple knock-out immunocompromised mice. Prostate. 2015;75:585–592. doi: 10.1002/pros.22939. [DOI] [PubMed] [Google Scholar]
- 33.Choi YY, Lee JE, Kim H, Sim MH, Kim KK, Lee G, Kim HI, An JY, Hyung WJ, Kim CB. Establishment and characterisation of patient-derived xenografts as paraclinical models for gastric cancer. Sci Rep. 2016;6:22172. doi: 10.1038/srep22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9:665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Table shows the panel of immunohistochemical markers analyzed and of the antibodies employed.
EBV score (Equivalent EBV Genomes/100000 cells) in primary gastric carcinomas and in the corresponding Lymphoma PDXs originated in NOD/SCID mice.
Supplementary Materials and Methods