Key Points
The Ikaros/IRF4 complex represses Ebf1 expression by binding to composite elements within the locus.
Ezh2 expression is induced by the Batf/IRF4 complex and the Ebf1-Pax5-Bach2 axis.
Abstract
The transcription factor (TF) interferon regulatory factor-4 (IRF4) promotes both germinal center (GC) reactions and plasma cell (PC) differentiation by binding to alternative DNA motifs including AP-1-IRF composite elements, Ets-IRF composite elements (EICEs), and interferon sequence response elements (ISREs). Although all of these motifs mediate transcriptional activation by IRF4, it is still unknown how some of the IRF4 target genes are downregulated upon PC differentiation. Here, we revealed a molecular mechanism of IRF4-mediated gene downregulation during PC differentiation. By combining IRF4 chromatin immunoprecipitation sequence and gene expression analysis, we identified zinc finger–IRF composite elements (ZICEs) in IRF4 binding regions aligned with genes whose expression was downregulated in PCs. The zinc finger TFs Ikaros and Aiolos were identified as IRF4 binding partners in PCs, and Ikaros but not Aiolos was essential for IRF4 binding to the ZICE sequence and for PC differentiation. The Ebf1 gene, which positively controls B-cell activation and GC reactions, was identified as one of the Ikaros/IRF4 target genes. Importantly, while the ZICE embeds the ISRE motif, IRF4 bound the ZICE motif as heterodimers with Ikaros for repression of target genes, which include Ebf1. In contrast, if the zinc finger motif is juxtaposed to the EICE motif, the Ikaros/PU.1/IRF4 complex functioned to activate target gene expression. Our findings revealed a novel mode of IRF4 activity upon PC differentiation where upon forming an Ikaros/IRF4 DNA-bound complex, a subset of genes is repressed.
Visual Abstract
Introduction
Cell differentiation is orchestrated by gene regulatory networks (GRNs) in which a network of transcription factors (TFs) coordinates the expression of cell-specific genes as the architectural genes.1,2 B cells are unique in that they undergo somatic cell genome modifications to diversify antibody function by class switch recombination (CSR) and somatic hypermutation of antibody genes when differentiated as germinal center (GC) B cells.3 Several key TFs, such as IRF4, Batf, Bcl6, Pax5, Bach2, and Blimp-1, have been shown to constitute the GRN specifying GC B-cell and plasma cell (PC) differentiation.4,5 The induction of Aicda gene encoding activation-induced cytidine deaminase is essential for CSR, and IRF4, Batf, and Pax5 have been shown as inducers of Aicda expression.6-9 Bach2 is required for CSR by repressing Prdm1 gene encoding Blimp-1 cooperatively with Bcl6.10-12 A positive feedback loop of IRF4–Blimp-1 drives terminal differentiation of activated B cells to PCs.13,14 Thus, the regulatory interaction of TFs organizes GC reaction during the course of PC differentiation.
IRF4 is essential for the expression of both GC B-cell–specific and PC-specific genes. Such diverse functions of IRF4 are thought to rely on multiple DNA binding motifs to which IRF4 binds as heterodimers or its homodimer depending on its protein level.15 IRF4 binds the Ets-IRF composite elements (EICEs) with PU.1,16 the AP-1-IRF composite elements (AICEs) with AP-1 family such as Batf,17-20 and the interferon sequence response elements (ISREs) as a homodimer. Each motif uniquely activates the expression of genes related to GC B-cell or PC differentiation.15 In particular, when its protein amount is low, IRF4 predominantly occupies AICE and EICE motifs on IRF4 target genes and contributes to Aicda expression, as well as the activation of Bcl6, specifying the functions of GC B cell.15,21 When its protein amount increases during PC differentiation, IRF4 binds the ISREs of direct target genes such as Prdm1. IRF4 levels thereby mediates cell fate decisions by coordinating its binding partner- and DNA-binding activity.
Besides gene activation, IRF4 has also been implicated in gene downregulation.15 A previous gene expression analysis of IRF4 direct target genes during PC differentiation revealed the presence of 3 major clusters; upregulated genes when IRF4 amount is low, and up- or downregulated genes when IRF4 amount is high.15 However, the molecular mechanism of IRF4-mediated gene downregulation has not been elucidated. Another important question is the regulation of Ezh2 in GC B cells. Ezh2, a subunit of polycomb repressive complex 2 (PRC2), maintains lower Irf4 expression.22,23 Because of the function, Ezh2 is required for GC B cells, and a reduced function of Ezh2 appears to promote PC differentiation. Therefore, the regulation of Ezh2 expression is an important matter to be elucidated. In this study, we address how a subset of IRF4 target genes is transiently induced or downregulated during PC differentiation. We found a DNA sequence-specific interaction between IRF4 and the zinc finger TF Ikaros. In addition to the role of Ikaros as a critical regulator of early lymphoid cell development, we here propose that Ikaros modulates the function of IRF4 during PC differentiation.
Methods
Full details on RNA sequencing (RNA-seq), chromatin immunoprecipitation (ChIP) assay, reverse transcription polymerase chain reaction (RT-PCR), complex purification and liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, immunoblot analysis and immunoprecipitation, flow cytometry, retroviral vectors and transduction of naïve B cells, stealth RNA interference, luciferase assay, electromobility shift assay, oligonucleotide precipitation assay, and statistical analysis are provided in supplemental Methods.
Mice
The B1-8hi gene targeted mice have been described24 and obtained from T. Kurosaki and M. Nussenzweig. Irf4-deficient mice have been described.25 Mice were maintained in pathogen-free conditions in accordance with guidelines approved by the institutional review boards of Tohoku University (2016MdA-215) and Nagasaki University (1503171206-2).
Cell culture
Splenic B cells were isolated as described.15 Cells were plated into culture at 0.5 × 106 to 1.0 × 106 cells per mL and stimulated with recombinant mouse interleukin-2 (IL-2; 100 U/mL), recombinant mouse IL-4 (5 ng/mL), recombinant mouse IL-5 (1.5 ng/mL), recombinant mouse CD40L (0.2 ng/mL) (all R&D Systems) for B1-8hi derived splenic B cells, or IL-4 (10 ng/mL) and CD40L (100 ng/mL) for wild-type (WT) and Irf4-deficient mice derived splenic B cells. NP(40)-ficoll (Biosearch Technologies Inc.) was used at 0.01 ng/mL concentration.
Results
IRF4 regulates 3 distinct gene subprograms during PC differentiation
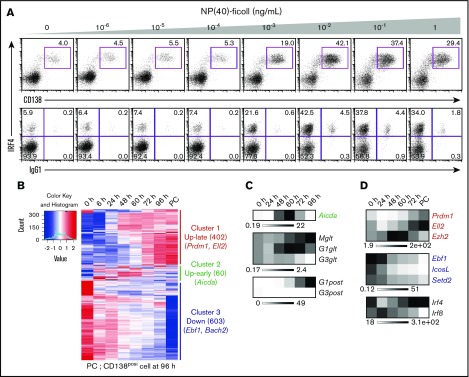
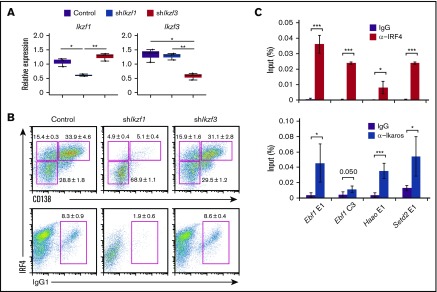
In this study, we used splenic B cells from B1-8hi B-cell antigen receptor (BCR) heavy chain knock-in mice,24 which have heavy chain loci targeted with high-affinity B1-8VH gene. When B1-8hi is combined with immunoglobulin λ (Igλ) light chains, B cells respond to the hapten 4-hydroxy-3-nitrophenylacetyl (NP). By referring to previous study,21 we first set up an efficient ex vivo PC differentiation system. By modulating BCR strength with different concentrations of NP-ficoll, we found a condition (10−2 ng/mL NP-ficoll) in which CD138 positive (CD138posi) PCs were detected at nearly 50% and were accompanied with class switch into IgG1 (Figure 1A). Under this condition, a time course transcriptome analysis discovered 3 major gene clusters based on their expression profiles; cluster 1 (up-late), cluster 2 (up-early), and cluster 3 (down) (Figure 1B). Genes that belong to cluster 2 showed a transient induction upon activation. We found Prdm1 and Ell2 in cluster 1, Aicda in cluster 2, and Ebf1 and Bach2 in cluster 3, patterns consistent with their functions (Figure 1B). Germline transcripts of IgG1 (G1glt) were increased from 24 hours after stimulation, and postswitch transcripts of IgG1 (G1post) were detected following the transient induction of Aicda expression from 48 to 60 hours (Figure 1C), suggesting that CSR occurred around these time points. Thus, the expression of these genes in B1-8hi splenic B cells after differentiation stimuli showed patterns consistent with the present model (supplemental Figure 1A).
Figure 1.
Ex vivo PC differentiation system using B1-8hisplenic B cells. (A) The dynamics of IRF4 expression and class-switched IgG1 frequency in response to differing BCR signaling intensities. Splenic B cells were purified from B1-8hi mice and stimulated with IL-2, 4, and 5 and CD40L with indicated concentrations of NP(40)-ficoll. IRF4 expression was analyzed at 72 hours with costaining of CD138 and IgG1. Data are representative of 2 independent experiments. (B) Heat map showing alteration of gene expression along PC differentiation. B1-8hi splenic B cells were isolated and stimulated with IL-2, 4, and 5, CD40L, and 10−2 ng/mL of NP(40)-ficoll, and messenger RNA (mRNA) was extracted at indicated time followed by RNA-seq. Cluster 1 (up-late), genes upregulated along differentiation; cluster 2 (up-early), genes transiently upregulated around 60 hours; cluster 3 (down), genes downregulated along differentiation. Each cluster contains 402, 60, and 603 genes with indicated genes, respectively. (C) Heat maps showing transcripts of Aicda from panel B and RT-PCR of germ line and postswitched immunoglobulin gene. For RT-PCR, results are presented relative to the abundance of transcripts encoding β2-microglobulin (Β2m), and the average expression is from 1 experiment using 3 mice. (D) Heat maps showing transcripts of indicated IRF4 direct target genes with that of Irf4 and Irf8 from panel B. For panels B-D, genes are indicated with each color: red, cluster 1; green, cluster 2; blue, cluster 3, respectively.
To clarify the molecular mechanisms of promoting CSR and PC differentiation by IRF4 in our cell system, we extracted “direct target genes of IRF4” from each cluster in the transcriptome data (Figure 1B). Toward this end, we first identified direct target genes of IRF4 using previously reported data sets of IRF4 ChIP sequence (ChIP-seq) in B1-8i splenic B cells and microarray comparing WT vs Irf4-deficient B cells (supplemental Figure 1B).15 By comparing this curated set of IRF4 target genes with our transcriptome measurements (402 up-late regulated genes [cluster 1] and 603 downregulated genes [cluster 3]), we found 96 and 78 genes, respectively, which we infer are bound and regulated by IRF4 (supplemental Table 1; supplemental Figure 2A-G). Prdm1, Ell2, and Ezh2 were activated by IRF4, whereas Ebf1, IcosL, and Setd2 were supposed to be downregulated by IRF4 (Figure 1D). IcosL regulates the interaction of B and T cells in GC,26 whereas Setd2 promotes Aicda transcription.27 Importantly, some of IRF4 binding regions surrounding these gene loci contain both AICE and ISRE motifs (supplemental Table 2). IRF4 physically interacts with Batf on the AICE sequence, and the binding of IRF4 and Batf was detected at Ell2 C1 and Ezh2 E1 regions (supplemental Figure 2H,I). Sequence specific binding of IRF4 to an ISRE motif at the Ezh2 E1 was confirmed by electrophoretic mobility shift assays (EMSAs) (supplemental Figure 2J). Among 60 up-early regulated genes (Figure 1B, cluster 2), Aicda was found as a direct target of IRF4, and IRF4 and Batf cooperatively bound to their regulatory regions at the Aicda locus (supplemental Tables 1 and 2; supplemental Figure 2K-M). To confirm IRF4-dependent regulation of these putative target genes, retroviral transduction of IRF4 in Irf4-deficient splenic B cells was performed. IRF4 complementation rescued the expression of these genes as well as PC differentiation (supplemental Figure 3). Therefore, IRF4 directly regulates 3 subprograms of gene expression upon PC differentiation that includes Ezh2.
Identification of zinc finger–IRF composite elements
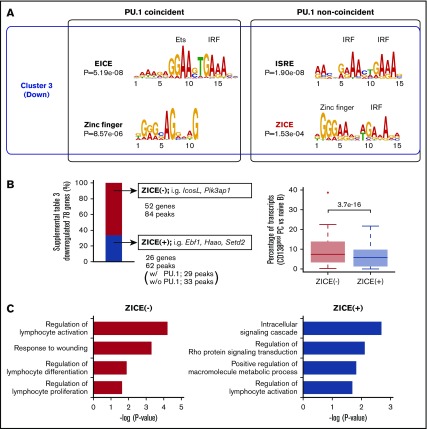
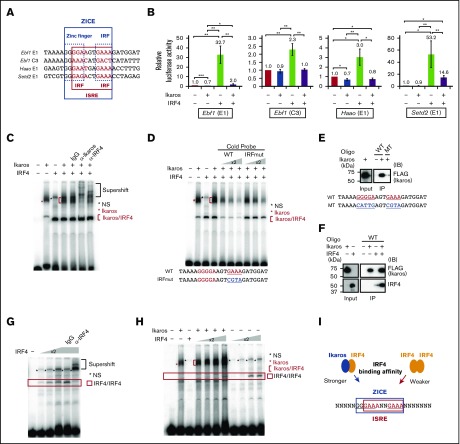
To identify regulatory motifs that could correlate with IRF4-dependent gene repression, DNA sequences from IRF4 binding regions of cluster 1 and 3 genes were compared by Multiple EM for Motif Elicitation (MEME). Consistent with our previous report,15 IRF4 binding peaks of cluster 1 genes contained EICE motifs as well as PU.1 ChIP-seq peaks, and AICE and ISRE motifs without PU.1 ChIP-seq peaks (supplemental Figure 4). In contrast to cluster 1, IRF4 bound regions of cluster 3 genes contained EICE and zinc finger motifs as well as PU.1 ChIP-seq peaks (Figure 2A). This observation was consistent with a previous report that found Ikaros colocalized with PU.1 and IRF4 binding regions in Rag2−/− pro-B cells.28 Through further analysis of these sequences, we identified a new motif, named zinc finger–IRF composite elements (ZICEs), within IRF4 peaks that did not overlap with PU.1 ChIP-seq peaks. The ZICE is composed of the zinc finger motif (GGGAA) and the IRF motif (GAAA) with a 3-nucleotide insertion between them.
Figure 2.
The newly identified ZICEs correlate with gene downregulation. (A) Motif analysis of IRF4 binding sequences obtained from downregulation target genes. IRF4 direct targets in B1-8hi splenic B cells were selected using IRF4 ChIP-seq in B1-8i splenic B cells (Gene Expression Omnibus accession number GSE46607) as described in supplemental Figure 1B. IRF4 binding targets belonging to cluster 3 (supplemental Table 1) were further classified into those with and without PU.1 binding (PU.1 coincident and noncoincident, respectively). In total, 47, 28, 113, and 47 regions were extracted for these categories, respectively. These sequences were analyzed with the MEME algorithm to identify overrepresented motifs within 100 bp in either direction for PU.1 coincident or 200 bp in either direction for PU.1 noncoincident of the peak maxima. Results are represented for the enriched motifs. (B) The ZICEs correlate with efficient downregulation of IRF4 direct target genes. Left: IRF4 downregulated target genes were classified into 2 subgroups depending on the presence of the ZICEs within the IRF4 bound regions (supplemental Table 3). ZICE (−), 52 genes with 84 IRF4 binding peaks that lacked the ZICEs and included IcosL and Pik3ap1; ZICE (+), 26 genes with 62 IRF4 binding peaks that contained the ZICEs and included Ebf1, Haao, and Setd2. Among these 62 peaks, 29 peaks were detected in PU.1 ChIP-seq as well, whereas 33 peaks were not detected. The y-axis shows the percentage of downregulated genes. Right: The amounts of transcripts of each ZICE (−) or ZICE (+) IRF4 direct target genes in CD138 positive (CD138posi) cells were divided by that of transcripts at 0 hours (from Figure 1B). Data are shown with box-and-whisker plot and the P value. (C) Gene ontology (GO) analysis of subgroups of IRF4 downregulation target genes. 52 ZICE (−) genes or 26 ZICE (+) genes were analyzed for their enrichment in GO focusing on biological process using the David v6.8 algorithm. The x-axis shows the P value of pathway-specific enrichment.
To examine functional activity of the ZICEs, IRF4 downregulated target genes were classified into 2 subgroups depending on the presence of the ZICEs within the IRF4 bound regions (supplemental Table 3). Fifty-two genes lacked the ZICEs and included the IcosL and Pik3ap1 loci, whereas 26 genes contained the ZICEs and included the Ebf1, Haao, and Setd2 loci (Figure 2B, left). We found that genes containing the ZICEs exhibited substantially more repression than those genes that lacked the ZICEs in our transcriptome measurement (Figure 2B, right). Gene ontology analysis indicated that both subclusters were enriched for genes involved in lymphocyte activation (Figure 2C). Uniquely, lymphocyte differentiation and proliferation functioning genes were enriched in the subcluster that lacked the ZICEs, whereas genes involved in signaling were enriched in the subcluster that contained the ZICEs. These suggest that the ZICE motif coevolved with a unique subgene program that specifies PC fate and function.
Ebf1 collaboratively promotes CSR with the IRF4/Batf complex
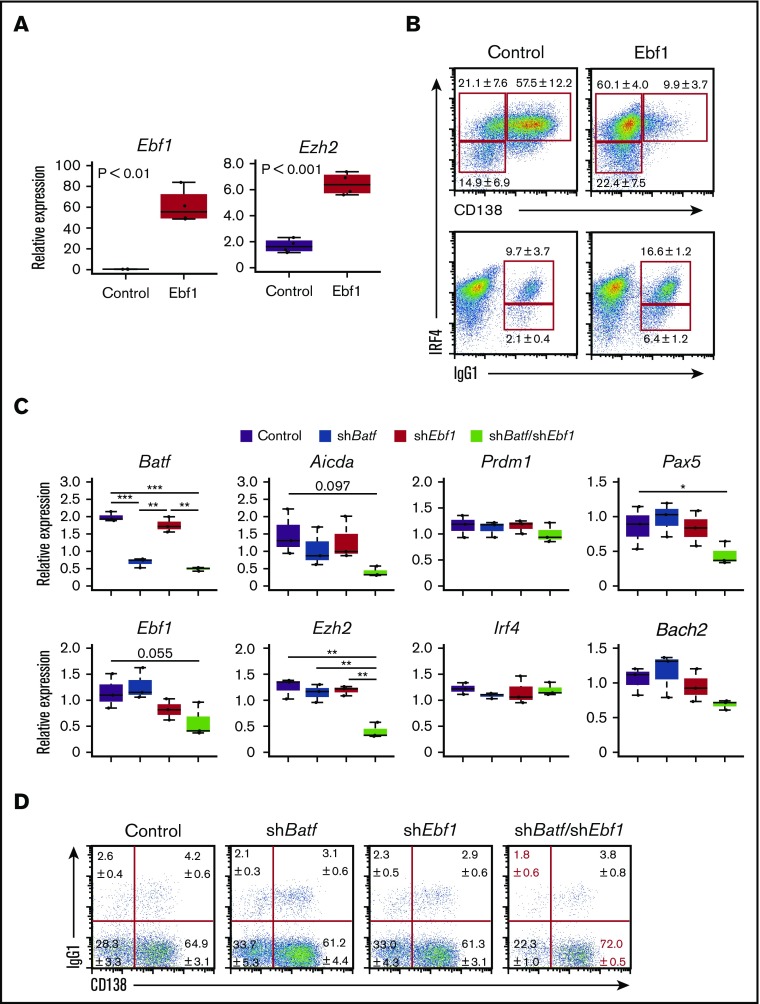
Among genes containing ZICEs, we focused on Ebf1, which is required for CSR by inducing G1glt and G3glt26 as well as repressing Prdm1 expression.29,30 We confirmed published observations of Ebf1 in our cell system by retrovirus transduction (Figure 3A). In Ebf1-transduced cells, the expression of TFs Pax5, target of Ebf1,31 and Bach2, target of Pax5,32 were induced (supplemental Figure 5A). In addition, Prdm1 expression was reduced, which may be a joint effect with Bach2.10 Aicda expression was induced, and G1glt showed greater expression. Furthermore, we found that Ezh2 expression was induced (Figure 3A). Given that Irf4 and Batf expression were not altered, it is possible that Ebf1 positively regulates Ezh2 expression in a Batf/IRF4 complex independent manner. At the cell level, the formation of CD138posi PCs was reduced and the number of IgG1posi cells was increased in Ebf1-transduced cells compared with control transductions (Figure 3B). Combined with previous reports,11,29,30 these results suggest that the Ebf1-Pax5-Bach2 axis promotes the expression of CSR-related genes and inhibits those involved in PC differentiation (supplemental Figure 5B).
Figure 3.
Ebf1 and the Batf/IRF4 complex cooperatively promote the expression of Aicda and Ezh2. (A-B) Ebf1 transduction in B1-8hi splenic B cells. Cells were transduced with control retroviral vector or retroviral vector expressing Ebf1 on day 1 after differentiation stimuli and sorted on the basis of green fluorescent protein (GFP) expression on day 3. One experiment was performed using 3 mice. (A) RT-PCR analysis of Ebf1 and Ezh2 transcripts. (B) Flow cytometry analysis of intracellular IRF4, CD138, and IgG1. Numbers adjacent to outlined areas indicate percent IRF4loCD138nega cells (top lower), IRF4hiCD138nega cells (top upper left), IRF4hiCD138posi cells (top upper right), IRF4loIgG1posi cells (bottom lower), or IRF4hiIgG1posi cells (bottom upper). (C-D) Knockdown of Batf and/or Ebf1 in B1-8hi splenic B cells. Cells were transduced with a control vector or vector targeting Batf (shBatf) and vector targeting Ebf1 (shEbf1) on day 1 after differentiation stimuli. On day 3, cells were sorted on the basis of GFP expression for control, and GFP (shBatf) and/or dsRed (shEbf1) expression for shBatf and/or shEbf1. One experiment was performed using 3 mice. (C) RT-PCR analysis of transcripts of indicated genes. (D) Flow cytometry analysis of surface CD138 and IgG1. For panels A and C, results are presented relative to the abundance of transcripts encoding Β2m, and shown with box-and-whisker plot. *P < .05; **P < .01; ***P < .001. For panels B and D, data are representative of 3 mice, and shown with the means and standard deviation (SD), respectively.
Because Aicda and Ezh2 are also regulated by the Batf/IRF4 complex, we examined the contribution of the Ebf1-Pax5-Bach2 axis to their regulation. Knockdown of Batf only marginally affected the expression of Aicda and Ezh2, whereas knockdown of Ebf1 had no effect (Figure 3C). In contrast, Aicda and Ezh2 expression was markedly reduced upon knockdown of both Batf and Ebf1. In these cells, Prdm1 and Irf4 expression was not altered, and Pax5 and Bach2 expression showed a marginal reduction. At the cell level, CD138posiIgG1nega cells were increased, and CD138negaIgG1posi cells were reduced (Figure 3D). Therefore, the Batf/IRF4 complex collaborates with the Ebf1-Pax5-Bach2 axis to promote CSR by activating Aicda and Ezh2 in these cells.
IRF4 interacts with the Ikaros-NuRD complex in B1-8hi splenic B cells
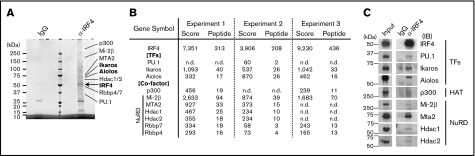
To identify the zinc finger TF that binds the ZICEs with IRF4, we purified the IRF4 complex in stimulated B1-8hi splenic B cells at day 3. Several unique bands were detected in IRF4 immunoprecipitates compared with that of control IgG. IRF4 complex components were identified using LC-MS/MS and confirmed by immunoblot analysis (Figure 4A-C). PU.1 was detected by LC-MS/MS in 1 of 3 experiments with low protein score and small numbers of peptides (Figure 4B). Upon differentiation, PU.1 expression was dramatically reduced at protein level (supplemental Figure 6A-B). Consistent with previous reports that PU.1 inhibits PC differentiation,15,33,34 PU.1 transduction blocked PC differentiation, and this was dependent on the DNA-binding activity of PU.1 as shown by transduction with a mutant PU.1 that disrupts the DNA binding domain (supplemental Figure 6C). Thus, the reduction of PU.1 appears prerequisite for PC differentiation. In regard to Batf, its expression was a transient and highest prior to PC differentiation (supplemental Figure 6A-B).
Figure 4.
Identification of Ikaros family proteins as IRF4 complex components in B1-8hisplenic B cells. B1-8hi splenic B cells were stimulated for 72 hours, and whole cell extracts were immunoprecipitated with control immunoglobulin (IgG) or anti-IRF4 (α-IRF4) antibodies. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis and MeOH-free Coomassie Brilliant Blue staining. IRF4 complex components were indicated at expected molecular weight. (B) LC-MS/MS analysis of the IRF4 complex. IRF4 complex components were determined as specific detection with α-IRF4, or more than twofold protein score with α-IRF4 than control IgG. Selected IRF4 complex components were shown with protein score and number of unique peptide from 3 independent experiments. (C) Immunoprecipitation followed by immunoblot analysis of indicated IRF4 complex components. Input, 2% of whole cell extracts. Data are representative of 1 of 3 (A) or 1 of 2 (C) independent experiments. n.d., not detected.
Among IRF4 complex components, we found the presence of p300 histone acetyltransferase suggesting that IRF4-mediated gene activation involves acetyl-dependent regulation. Importantly, we detected Ikaros and Aiolos in IRF4 immunoprecipitates (Figure 4A-C). Ikaros has been shown to interact with the nucleosome remodeling and deacetylase (NuRD) complex,35 whose components were also detected in the IRF4 complex. The protein score of Ikaros detection was two- to threefold higher than that of Aiolos in 2 of 3 experiments, suggesting that Ikaros is the dominant partner in the IRF4 complex we characterized. Of note, Ikaros expression was unchanged during PC differentiation (supplemental Figure 6A-B).
Ikaros and Aiolos are important for early lymphoid differentiation.36,37 Although Aiolos is required for long-lived PCs in the bone marrow,38 the role of Ikaros in PCs has not been reported. To examine their possible roles in PC differentiation, we performed knockdown experiments of Ikaros and Aiolos in our cell system. Importantly, knockdown of Ikaros markedly reduced PC differentiation (Figure 5A-B). In contrast, knockdown of Aiolos displayed marginal effects on PC differentiation. The binding of IRF4 and Ikaros to selected regions harboring ZICE sequences was confirmed with ChIP–quantitative PCR (Figure 5C). These results suggest that Ikaros is essential for PC differentiation and that Ikaros could bind the ZICEs with IRF4.
Figure 5.
Knockdown of Ikaros but not Aiolos resulted in defect of plasma cell differentiation. (A-B) Knockdown of Ikaros or Aiolos in B1-8hi splenic B cells. Cells were transduced with a control vector or vector targeting Ikaros (shIkzf1) or Aiolos (shIkzf3) on day 1 after differentiation stimuli, then sorted on the basis of GFP expression on day 3. One experiment using 3 mice. (A) RT-PCR analysis of Ikzf1 and Ikzf3 transcripts. Results are presented relative to the abundance of transcripts encoding Β2m, and shown with box-and-whisker plot. (B) Flow cytometry analysis of intracellular IRF4 and CD138 and IgG1. Numbers adjacent to outlined areas indicate percent IRF4loCD138nega cells (top lower), IRF4hiCD138nega cells (top upper left), IRF4hiCD138posi cells (top upper right), or IgG1posi cells (bottom). Data are representative of 3 mice, and shown with the mean and SD, respectively. (C) The binding of IRF4 or Ikaros to their regulatory regions at the Ebf1, Haao, and Setd2 loci. B1-8hi splenic B cells were stimulated for 48 hours, and ChIP assay was performed using control IgG, α-IRF4, or α-Ikaros, followed by quantitative PCR analysis. Binding enrichments is presented relative to input DNA. The average enrichment and SD is from 3 independent experiments for α-IRF4, and 2 independent experiments with technical duplicate for α-Ikaros. For panels A and C, *P < .05; **P < .01; ***P < .001.
Ikaros functions as an activator with PU.1
Prior to focusing on the ZICEs, we examined the role of a zinc finger motif detected along the EICE motif (Figure 2A). The binding of PU.1 or IRF4 to selected regions was confirmed by ChIP–quantitative PCR, and we found that Ikaros was recruited to them as well (supplemental Figure 6D-E). Although these observations suggest that Ikaros might modify EICE-mediated gene regulation by IRF4, the effect of a zinc finger motif juxtaposed to the EICE motif on gene expression has not been clear. Therefore, we performed luciferase assays using Ebf1 C4 and IcosL E2 regulatory regions, which naturally contain zinc finger motifs juxtaposed to EICE motifs (supplemental Figure 6D). As expected, reporter activity was activated by PU.1 transfection and was further enhanced by IRF4 cotransfection (supplemental Figure 6F). IRF4 had no activity on its own. Interestingly, Ikaros could potentiate PU.1-dependent reporter activity to the same extent as that observed with IRF4. Furthermore, cotransfection of the 3 TFs resulted in synergistic enhancement of reporter activity. However, Ikaros did not exhibit any activity on its own or with IRF4. Thus, these results indicated that the combination of EICE and zinc finger motifs promotes maximal activation of reporter activity. Importantly, these results suggest that Ikaros can function as a transcriptional activator depending on the context, in this case in the presence of PU.1.
Binding of Ikaros/IRF4 complexes to the ZICEs inhibits IRF4-mediated gene activation
Next, we examined the role of the ZICE sequence. We noticed that the zinc finger motif of the ZICEs includes an IRF motif (Figure 6A). We performed luciferase assays using Ebf1 E1 and C3, Haao E1, and Setd2 E1 regulatory regions, which naturally harbor ZICE sequences. Ikaros transfection slightly repressed or exhibited no effect on these reporters (Figure 6B). In contrast, IRF4 moderately or markedly activated the expression of the same reporters. Interestingly, cotransfection of Ikaros and IRF4 attenuated the ability of IRF4 to activate expression of these reporters. These results indicate that Ikaros attenuates IRF4-dependent gene activation and that this effect could be mediated by the ZICE sequence in these regulatory elements.
Figure 6.
Ikaros recruits IRF4 to the ZICEs and inhibits IRF4-mediated gene activation. (A) ZICE sequences of indicated regions. ZICE, GGGAANNNGAAA indicated with blue box; ISRE, GAAANNGAAA indicated with red box. (B) Luciferase assays using the ZICE containing reporter genes. 293T cells were transiently transfected with indicated reporter and effector plasmids. The amounts of plasmids were as follows: luciferase reporter (1.0 μg), Ikaros (100 ng), and IRF4 (100 ng). The reporters used are shown above each panel. The average luciferase activity and SD are from 3 independent experiments. *P < .05; **P < .01; ***P < .001. (C) Binding of Ikaros/IRF4 complexes to the ZICE motif. EMSA with the Ebf1 E1 sequence as probe. α-Ikaros or α-IRF4 or control antibodies were used in supershift assays to confirm Ikaros/IRF4 complexes. (D) Competition assay using the Ebf1 E1 probe. Increased amounts of competitor DNAs, WT or mutant IRF (IRFmut), were included as indicated. (E-F) Oligonucleotide precipitation assay using the Ebf1 E1 oligonucleotide. Biotinylated WT or mutant (MT) oligonucleotide was incubated with nuclear extracts as indicated. MT carries mutation in both a zinc finger and an IRF motifs. Oligonucleotide-protein complex was immunoprecipitated with streptavidin beads followed by immunoblot. As input, 10% of immunoprecipitation (IP) for Ikaros or 50% of IP for IRF4 were loaded. Immunoblot was performed using α-FLAG (M2) for detecting Ikaros or α-IRF4. (G) Binding of IRF4 homodimer to an ISRE motif within the ZICE of Haao E1 probe. α-IRF4 or control antibodies were used in supershift assays to confirm IRF4/IRF4 complexes. (H) The effective recruitment of IRF4 to the ZICE sequence in the presence of Ikaros rather than to the ISRE sequence. All binding reactions contain a Haao E1 probe. IRF4 concentration was increased in twofold increments as indicated in the presence or absence of Ikaros. For (C-H), nuclear extracts were prepared from 293T cells transfected with pcDNA3 HA-IRF4 or Flag-Ikaros expressing vector, respectively. Red asterisk, Ikaros specific band; red bracket, Ikaros/IRF4 complexes; red box, IRF4/IRF4 complexes; black asterisk, nonspecific (NS). (I) Schematic representation of IRF4 recruitment to the ZICEs. ZICE, GGGAANNNGAAA underlined and indicated with blue box; ISRE, GAAANNGAAA indicated with red box. The ZICEs embed the ISRE motif, and IRF4 enables to bind the ZICE sequence as a heterodimer with Ikaros or the ISRE sequence as a homodimer. However, IRF4 is effectively recruited to the ZICE sequence in the presence of Ikaros with lower IRF4 concentration. Therefore, the Ikaros/IRF4 complex binds the ZICEs for repressing target genes.
To establish the Ikaros/IRF4 interaction and their assembly on ZICE sequences, we performed EMSAs using nuclear extracts from 293T cells transfected with individual protein expression vectors. Ikaros generated a protein-DNA complex with a probe containing ZICE sequence, whereas IRF4 or Aiolos did not (Figure 6C; supplemental Figure 7A). When Ikaros and IRF4 were combined, the intensity of the specific band became stronger and broader. The band was supershifted by adding reactive antibodies for Ikaros or IRF4, indicating that it contained both Ikaros and IRF4 (Figure 6C; supplemental Figure 7B). Sequence specificity of the Ikaros/IRF4 complex was examined by competition assay using cold probes. The complex was disappeared with increasing amounts of WT probe, and it still remained with same amounts of mutant probe (IRFmut), which carries mutation in the IRF motif. Although EMSAs suggested that the Ikaros/IRF4 complex assembles on ZICE sequences, it was still ambiguous. Therefore, we further performed an oligonucleotide precipitation assay. Ikaros was immunoprecipitated with WT oligo but not with mutant oligo, which carries mutation in both zinc finger and IRF motifs (Figure 6E). Importantly, IRF4 was efficiently immunopreciptated with WT oligo in the presence of Ikaros (Figure 6F). Combined with EMSAs, these results demonstrate that Ikaros/IRF4 coassembles on ZICE sequences, and that Ikaros is required to recruit IRF4.
To gain further insight on the binding properties of Ikaros and IRF4 to the ZICE sequence, we compared the binding kinetics over a wide range of IRF4 concentrations. IRF4 could bind on its own to a probe containing the ZICE sequence (Figure 6G). However, relatively high concentrations of IRF4 were required for binding (Figure 6H). In contrast, the complex observed in the presence of both Ikaros and IRF4 bound efficiently to the ZICE probe, even at low concentrations of IRF4. These results indicate that IRF4 is more efficiently recruited to ZICE sequences in the presence of Ikaros than to ISRE sequences (Figure 6I). Therefore, we propose that the inhibition of IRF4-mediated gene activation in the presence of Ikaros is caused, at least in part, by modulating IRF4 homodimer binding to the ISRE motif adjacent the ZICEs.
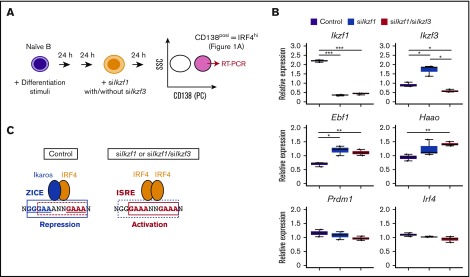
IRF4 is thought to occupy ISRE motifs with its high concentration for gene activation,15 and PCs are characterized as IRF4hi status7 (Figure 1A). To examine the functional importance of the ZICE, we performed knockdown of Ikaros with/without Aiolos in PCs. B1-8hi splenic B cells activated ex vivo were transfected with oligo duplex siRNA at 48 hours (Figure 7A). After another 24 hours, CD138posi PCs were sorted for quantitative gene expression analysis. Compared with control cells, Ikzf1 expression was reduced >80% in siIkzf1- or siIkzf1/siIkzf3-transfected cells (Figure 7B). It should be noted that Ikzf3 expression was induced approximately twofold in siIkzf1-transfected cells, and it was reduced ∼40% in siIkzf1/siIkzf3-transfected cells. Importantly, the expression of Ebf1 and Haao genes, which possess ZICE motifs within their loci, were upregulated in siIkzf1- or siIkzf1/siIkzf3-transfected cells. In contrast, the expression of Prdm1, which possesses an ISRE motif within the locus,15 or Irf4 was unaffected by these treatments. These observations suggest that ZICE motifs in the regulatory regions of the Ebf1 and Haao loci were converted to ISRE motifs with loss of Ikaros but not Aiolos in PCs (Figure 7C).
Figure 7.
Loss of Ikaros resulted in upregulation of Ebf1 and Haao genes in CD138posiplasma cells. (A) Schematic representation of knockdown in CD138posi PCs derived by differential stimulation. B1-8hi splenic B cells were stimulated for 48 hours and transfected with knockdown sequences targeting control or Ikaros (siIkzf1) with/without Aiolos (siIkzf3). Cells were cultured for another 24 hours, and CD138posi PCs were sorted and carried for RT-PCR in panel B. As shown in Figure 1A, CD138posi PCs correspond to IRF4hi fraction. (B) RT-PCR of transcripts of indicated genes in control cells and siIkzf1- or siIkzf1/siIkzf3-transfected CD138posi PCs. Ebf1 and Haao, shown as IRF4 target genes possessing ZICE sequences that embed ISRE motifs bound by IRF4 homodimer for gene activation. Prdm1, shown as an IRF4 target gene possessing an ISRE motif. Results are presented relative to the abundance of transcripts encoding Β2m, and shown with box-and-whisker plot. One experiment was performed using 3 mice. *P < .05; **P < .01; ***P < .001. (C) Schematic representation of the regulation of ZICE target genes, Ebf1 and Haao, in control and siIkzf1-or siIkzf1/Ikzf3-transfected CD138posi PCs. In control cells, IRF4 effectively binds the ZICE motif as a heterodimer with Ikaros for repressing target gene. In siIkzf1- or siIkzf1/siIkzf3-transfected cells, IRF4 binds the ISRE motif within the ZICEs as a homodimer, resulting in activation of these target genes. ZICE, GGGAANNNGAAA underlined and indicated with blue box; ISRE, GAAANNGAAA indicated with red box.
Discussion
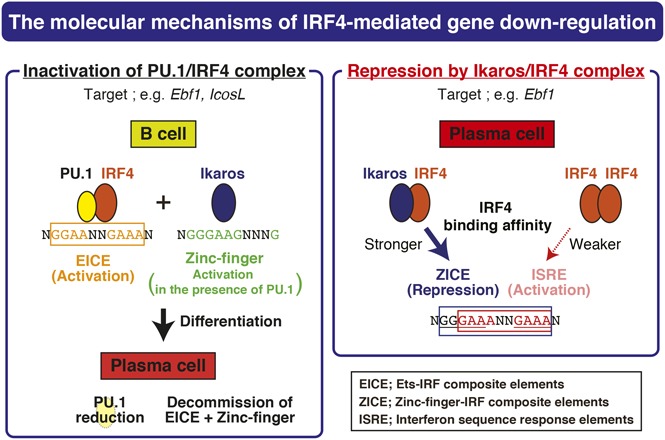
In this study, we revealed a mechanism of IRF4-dependent gene repression during PC differentiation. Importantly, the ZICEs were identified as a new motif shared among a subset of IRF4 target genes whose expression is reduced upon PC differentiation (Figure 2A). We provide the evidence that the Ikaros/IRF4 complex is bound to this motif (Figure 6C,F). Interestingly, the ZICEs embed the ISRE motif (Figure 6A) and were bound by IRF4 homodimer as well (Figure 6G). Because PCs are characterized as IRF4hi status in which IRF4 binds the ISREs,7 there is a possibility that the ISRE motif adjacent to the ZICEs mediates gene activation in PCs. However, IRF4 is more efficiently recruited to ZICE motifs in the presence of Ikaros (Figure 6H), resulting in the Ikaros/IRF4-mediated gene repression (Figure 6B,I). Consistent with this model, IRF4 target genes that harbor the ZICEs showed lower expression in PCs than those that lack the ZICEs (Figure 2B). Considering that IRF4 interacts with the Ikaros-NuRD complex (Figure 4), the molecular mechanism of Ikaros/IRF4-mediated gene repression likely involves epigenetic deacetylation of histones. Although we also detected Aiolos in the IRF4 complex, we could not detect binding of Aiolos to ZICE motifs. The regulatory motif of IRF4 overlaps with that of Blimp-1, and Aiolos contributes to Blimp-1 function.39 Thus, each Ikaros family member likely involves nonoverlapping roles in IRF4 and Blimp-1–dependent PC differentiation.
Our findings extend the IRF4 GRN that orchestrates CSR and PC differentiation (supplemental Figure 8). Although IRF4 is required for both GC B cells and PCs, its expression is kept at a relatively lower amount in GC B cells compared with PCs. IRF4lo status is essential for GC B cells because IRF4hi status prevents GC reactions including CSR.7 GC B cells maintain low levels of IRF4 in part by the actions of Ezh2.22,23 Ezh2 expression is robustly induced in GC B cells and reduced upon PC differentiation.23 Here, we have shown that Batf/IRF4 with Ebf1 functions to induce Ezh2 expression and impede PC differentiation (Figure 3A,C). GC B cells are distinguished into 2 statuses, the dark zone and the light zone. Batf expression is highly induced in the light zone B cells,40 and IRF4 is expressed at lower levels in GC B cells.7 Bach2 represses Prdm1 expression with Bcl6 in dark zone B cells,12,41 suggesting that the Ebf1-Pax5-Bach2 axis is activated in the dark zone B cells. Because activated B cells have not been separated into the light zone or the dark zone B cells in this study, we assume that our observations are of mixed cell status. Because of this, Ezh2 expression, as well as Aicda expression, might be effectively reduced by knockdown of both Batf and Ebf1 (Figure 3C). Therefore, we suggest a new feedback regulation between IRF4 and Ezh2 (supplemental Figure 8, upper).
In addition, we propose a distinct regulatory loop involving Ebf1 and IRF4. Our new results suggest that Ebf1 expression is also organized by at least 2 distinct manners, activation and repression by IRF4. The former is achieved by the PU.1/IRF4 complex and is further enhanced by Ikaros (supplemental Figure 8, upper), whereas the latter involves the Ikaros/IRF4 complex (supplemental Figure 8, lower). In both cases, Ikaros seems important. Knockdown of Ikaros in activated B cells resulted in a prominent blockage of PC differentiation, which is consistent with Irf4-deficient B cells (supplemental Figure 3A). Cooperative activities of PU.1, IRF4, and Ikaros could be required for initiating differentiation by activation of Ebf1 expression in GC B cells. We should note that the ZICE motif of Ebf1 E1 is overlapped with an EICE motif (Figure 6A). PU.1 binding to the region was detected in day-1 ChIP-seq, whereas it became obviously low in day-3 ChIP-seq (supplemental Figure 9A). Compared with PU.1 ChIP-seq, IRF4 binding to Ebf1 E1 and Haao E1 regions was consistently detected in day-1 and day-3 ChIP-seq (supplemental Figure 9A-C), and Ikaros bound these regions as well (Figure 5C). Considering the reduction of PU.1 amounts upon differentiation, we propose that Ikaros/IRF4 complexes replace PU.1/IRF4 complexes at EICE/ZICE overlapping motifs in PCs. In addition to Ebf1, some of Ikaros/IRF4 bound target genes are also regulated by PU.1/IRF4 (supplemental Table 3). Compared with the effect of gene activation by PU.1/IRF4, that of gene repression by Ikaros/IRF4 is weaker. PU.1 reduction is necessary to reveal IRF4-mediated gene downregulation at both ZICEs or EICEs and zinc finger motif regulatory regions. Together, IRF4 activity appears to be modified by the nature of partner TFs, PU.1, Batf, and Ikaros for orchestrating GC B-cell and PC differentiation.
Combined with previous reports, we expand the principal GRN orchestrating GC B-cell and PC differentiation. Now, an emerging question is how the balance of key TFs is maintained in GC B cells and then shifted to terminal differentiation. It is possible that changes of 1 or few of TFs in their amounts switch on balance shift. For example, Bach2 regulates the probability of undergoing CSR upon antigen stimuli42; however, its expression and protein stability is negatively regulated under BCR signaling.43,44 Thus, a reduction in Bach2 activity may trigger the GRN to switch from GC B cells to PCs. Although PU.1 expression decreases during differentiation, little is known about the molecular mechanisms governing the IRF4 and its other partner TFs, which could be regulated by ligation of cell surface receptors and signaling cascade. Consequently, understanding the regulation of these TFs by signaling cascades will provide clues to solve the transition of GRN from GC B cells to PCs.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank R. Sciammas (University of California, Davis), S. Tashiro (Hiroshima University), H. Singh (Cincinnati Children’s Hospital Medical Center), T. Ikura (Kyoto University), and D. Kurotaki (Yokohama City University) for helpful discussion; T. Kurosaki (Osaka University) and M. Nussenzweig (Rockefeller University) for providing B1-8hi mice; and M. Tsuda (Tohoku University) for operating the Illumina GAIIx.
This work was supported by Grants-in-Aid (JP24790271, JP25118701, JP16H01295, and JP16K19026) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Agency for Medical Research and Development–Core Research for Evolutional Science and Technology, Japan Science and Technology Agency. K.O. was supported by Takeda Science Foundation, the Naito Foundation, and the Uehara Memorial Foundation. Part of this study was supported by Biomedical Research Core of Tohoku University School of Medicine.
Authorship
Contribution: K.O. and K.I. conceived the project; K.O. designed and performed the majority of the experiments; H.K. assisted with luciferase assays; Y.O., K. Kinoshita, and T.N. analyzed RNA-seq data; Y.K. and H.S. performed LC-MS/MS analysis; K. Kimura and K.Y. assisted with Irf4-deficient mice experiments; R.F. and K.N. managed the Illumina GAIIx; K.O. wrote the manuscript; and K.I. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kyoko Ochiai, Department of Biochemistry, Tohoku University Graduate School of Medicine, Seiryo-machi 2-1, Sendai 980-8575, Japan; e-mail: kochiai@med.tohoku.ac.jp.
References
- 1.Singh H. Transcriptional and epigenetic networks orchestrating immune cell development and function. Immunol Rev. 2014;261(1):5-8. [DOI] [PubMed] [Google Scholar]
- 2.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613-626. [DOI] [PubMed] [Google Scholar]
- 3.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12(7):517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15(3):160-171. [DOI] [PubMed] [Google Scholar]
- 5.Shi W, Liao Y, Willis SN, et al. . Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015;16(6):663-673. [DOI] [PubMed] [Google Scholar]
- 6.Ise W, Kohyama M, Schraml BU, et al. . The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12(6):536-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25(2):225-236. [DOI] [PubMed] [Google Scholar]
- 8.Nera KP, Kohonen P, Narvi E, et al. . Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24(3):283-293. [DOI] [PubMed] [Google Scholar]
- 9.Gonda H, Sugai M, Nambu Y, et al. . The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198(9):1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muto A, Tashiro S, Nakajima O, et al. . The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429(6991):566-571. [DOI] [PubMed] [Google Scholar]
- 11.Ochiai K, Katoh Y, Ikura T, et al. . Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281(50):38226-38234. [DOI] [PubMed] [Google Scholar]
- 12.Ochiai K, Muto A, Tanaka H, Takahashi S, Igarashi K. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int Immunol. 2008;20(3):453-460. [DOI] [PubMed] [Google Scholar]
- 13.Minnich M, Tagoh H, Bönelt P, et al. . Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17(3):331-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tellier J, Shi W, Minnich M, et al. . Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol. 2016;17(3):323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochiai K, Maienschein-Cline M, Simonetti G, et al. . Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38(5):918-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9(11):1377-1387. [DOI] [PubMed] [Google Scholar]
- 17.Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13(7):499-509. [DOI] [PubMed] [Google Scholar]
- 18.Li P, Spolski R, Liao W, et al. . BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490(7421):543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tussiwand R, Lee WL, Murphy TL, et al. . Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490(7421):502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnamoorthy V, Kannanganat S, Maienschein-Cline M, et al. . The IRF4 gene regulatory module functions as a read-write integrator to dynamically coordinate T helper cell fate. Immunity. 2017;47(3):481-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7(1):495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Béguelin W, Popovic R, Teater M, et al. . EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caganova M, Carrisi C, Varano G, et al. . Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123(12):5009-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Shih TA, Meffre E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3(6):570-575. [DOI] [PubMed] [Google Scholar]
- 25.Honma K, Udono H, Kohno T, et al. . Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS. Proc Natl Acad Sci USA. 2005;102(44):16001-16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, Xu H, Shih C, et al. . T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517(7533):214-218. [DOI] [PubMed] [Google Scholar]
- 27.Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. J Biol Chem. 2012;287(39):32415-32429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwickert TA, Tagoh H, Gültekin S, et al. . Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15(3):283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Györy I, Boller S, Nechanitzky R, et al. . Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 2012;26(7):668-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi H, Nakayama M, Takami Y, Kuribayashi F, Nakayama T. EBF1 acts as a powerful repressor of Blimp-1 gene expression in immature B cells. Biochem Biophys Res Commun. 2012;422(4):780-785. [DOI] [PubMed] [Google Scholar]
- 31.Roessler S, Györy I, Imhof S, et al. . Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27(2):579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McManus S, Ebert A, Salvagiotto G, et al. . The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30(12):2388-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carotta S, Willis SN, Hasbold J, et al. . The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014;211(11):2169-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18(4):977-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Jackson AF, Naito T, et al. . Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2011;13(1):86-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JH, Nichogiannopoulou A, Wu L, et al. . Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5(6):537-549. [DOI] [PubMed] [Google Scholar]
- 37.Wang JH, Avitahl N, Cariappa A, et al. . Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9(4):543-553. [DOI] [PubMed] [Google Scholar]
- 38.Cortés M, Georgopoulos K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J Exp Med. 2004;199(2):209-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung KH, Su ST, Chen CY, et al. . Aiolos collaborates with Blimp-1 to regulate the survival of multiple myeloma cells. Cell Death Differ. 2016;23(7):1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue T, Shinnakasu R, Ise W, Kawai C, Egawa T, Kurosaki T. The transcription factor Foxo1 controls germinal center B cell proliferation in response to T cell help. J Exp Med. 2017;214(4):1181-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, Geng H, Boss I, Wang L, Melnick A. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood. 2014;123(7):1012-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muto A, Ochiai K, Kimura Y, et al. . Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29(23):4048-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ando R, Shima H, Tamahara T, et al. . The transcription factor Bach2 is phosphorylated at multiple sites in murine B cells but a single site prevents its nuclear localization. J Biol Chem. 2016;291(4):1826-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamahara T, Ochiai K, Muto A, et al. . The mTOR-Bach2 cascade controls cell cycle and class switch recombination during B cell differentiation. Mol Cell Biol. 2017;37(24):e00418-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.