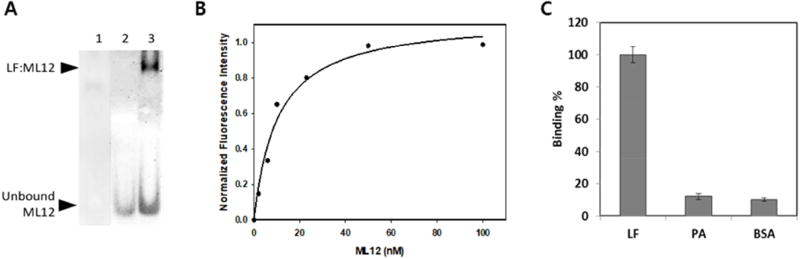
Figure 1.
Binding experiments between LF and ML12. (A) Gel shift assay between LF and the aptamer ML12. Lane 1 represents the gel result after loading only LF; Lane 2 represents the gel result after loading only ssDNAs; Lane 3 respresent the gel result after loading ssDNAs and LF. The top band shows the complex of LF and ML12 and the bottom band shows unbound ML12 in a 6 % native gel with 5.9 µM LF and 100 nM ML12 using SYBR gold staining. Prior to loading sample, 20 min incubation at 30 °C in buffer (30 mM Tris, pH 8.0, 100 mM NaCl, 5.0 mM KCl and 0.6 mM MgCl2) was conducted. The total sample volume was 10 µL. (B) Determination of binding affinity of aptamers. The black circles are for ML12 and the empty circles are for ML12 with flanking sequences. Binding reactions were carried out with a constant concentration of LF protein in a 96-well black plate and varied concentarions of Cy3-attached ML12. The fluorescent intensity of the bound ML12 or ML12 with flanking sequences to LF was measured after a couple of washing steps. The Kd value was determined by nonlinear fitting of the saturation binding curve. (C) Binding specificty of ML12 against BSA by the fluorescent intensity measurement of bound Cy3-ML12 to either LF or BSA. The concentration of the proteins (LF and BSA) was kept constant at 0.5 µM, and 10 nM of ML12 was used for initial incubation against the proteins.