Figure 2.
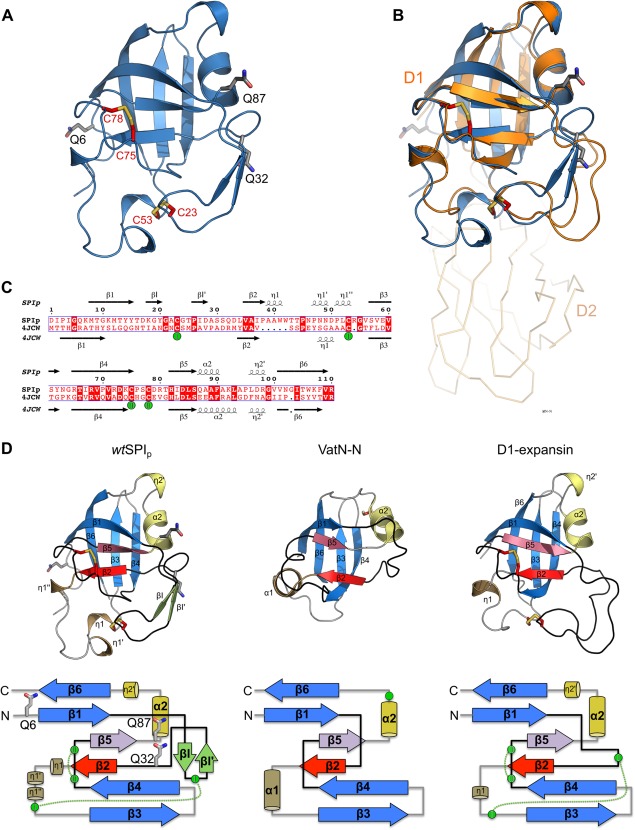
Crystal structure of the transglutaminase substrate (SPIp) from Streptomyces mobaraensis and tertiary structure comparison with DPBB‐containing proteins. (A) Cartoon representation of SPIp glutamines (carbons gray) and cysteines (carbons red) shown in stick representation. (B) Superposition of SPIp with the D1 N‐terminal domain of expansin (PDB 4JCW). SPIp is colored blue and expansin orange. The C‐terminal domain D2 of expansin is shown as ribbon representation. (C) Sequence alignment of SPIp and expansin D1 (PDB 4JCW) with secondary structure assigned by the DSSP algorithm (η indicates a 310‐helix). Disulfide‐bridges are numbered and cysteines involved are marked with green spheres. (D) Protein fold (cartoon representation) and topology comparison of SPIp (left), VatN‐N (middle, PDB 1CZ4), and the D1 domain of C. michiganensis’ expansin (right, PDB 4JCW). Each double psi‐beta‐barrel is composed of six β‐strands. β2 and β5, which are the central strands of each psi–loop (black), are colored red and violet, respectively. Remaining β‐strands that are part of the double‐psi‐beta barrel are highlighted in blue. For SPIp, two short β‐strands (βI and βI′) are inserted after β1 and are colored light green. Right‐handed α‐helices are colored sand (α1) and yellow (α2). 310‐helices (e.g., η1 or η2) are colored the same way. Cysteines in structures are shown as sticks with carbon atoms in red. Cysteines in the topology drawing are shown as green circles. Disulfide‐bridges are indicated by green dotted lines. Glutamine positions are shown as sticks with carbon atoms in gray for SPIp only.
