Figure 1.
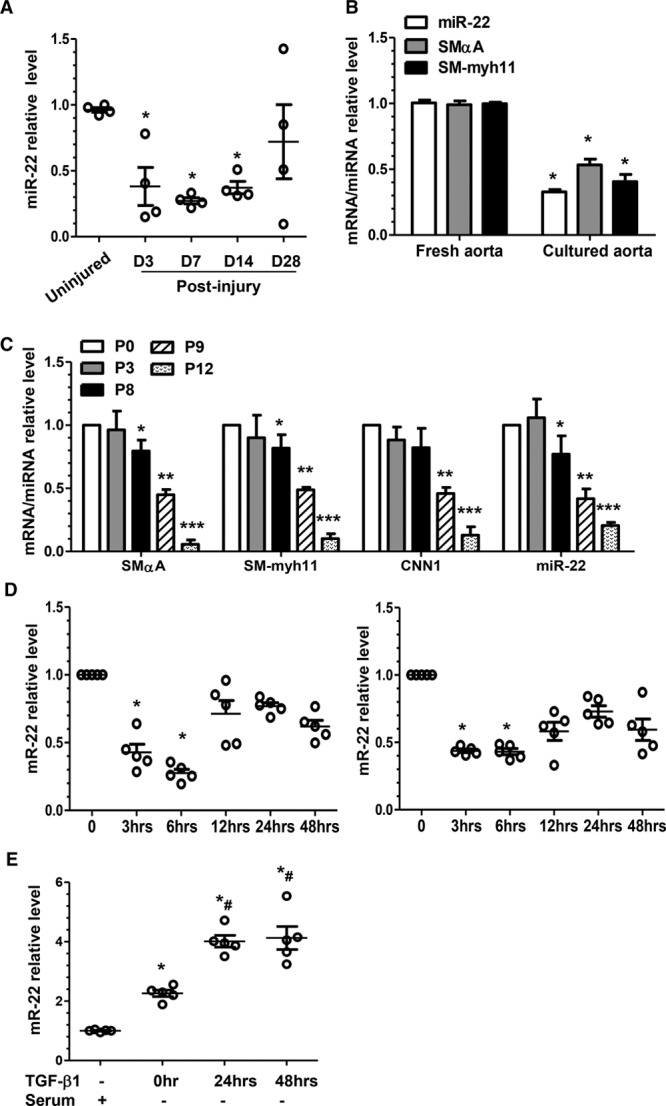
miR-22 expression is closely modulated during VSMC phenotype switching. A, miR-22 was significantly downregulated after injury in the in vivo mouse model. Total RNA was collected from the uninjured (day [D] 0) and injured femoral arteries (D3, D7, D14, and D28 postinjury). RT-qPCR analyses were conducted to obtain relative expression levels. B, miR-22 was decreased in the ex vivo cultured thoracic aortas. RT-qPCR analysis was used to examine the mRNA (SMαA and SM-myh11) and miR (miR-22) levels in the freshly isolated aortas and the aortas cultured in DMEM containing 20% serum for 3 days. Data and error bars in A and B represent the mean±SEM (n=4), where up to 5 femoral arteries were pooled as 1 experiment. *P<0.05 (versus D0 [A], or fresh aorta [B]). C, miR-22 expression was downregulated in the extended culture of murine VSMCs. Total RNA, including miR (miR-22) and mRNA (SMαA, SM-myh11, and CNN1), was harvested from freshly cultured VSMCs (cultured until day 7 and then split, designated P0), and VSMCs with the indicated passage number (P0, P3, P8, P9, or P12) were subjected to RT-qPCR analysis to obtain relative expression levels. D, Serum (left) and PDGF-BB (right) downregulated miR-22 in cultured VSMCs. VSMCs were subjected to serum starvation for 48 hours, followed by incubation with 20% serum or PDGF-BB (10 ng/mL), respectively. Total RNA was harvested at each indicated time point. E, Serum starvation and TGF-β1 upregulated miR-22 in cultured VSMCs. VSMCs in normal culture were used as control (TGF-β1–, Serum+) or subjected to serum starvation for 48 hours and then harvested after the indicated stimulations and time points: no stimulation (TGF-β1–, Serum–) at 0 hour, TGF-β1 stimulation at 24 hours (TGF-β1-24hrs, Serum–), and TGF-β1 stimulation at 48 hours (TGF-β1-48hrs, Serum–). Total RNA was extracted and subjected to RT-qPCR analysis with a specific miR-22 forward primer and a universal miR reverse primer. Data and error bars in C through E represent mean±SEM (n=5). *P<0.05, **P<0.01, ***P<0.001 (versus P0 [C], 0 hours [D], or normal culture [E]); #P<0.05 (versus 0 hours). DMEM indicates Dulbecco’s modified Eagle’s medium; miR-22, microRNA-22; PDGF-BB, platelet-derived growth factor BB; RT-qPCR, reverse transcription quantitative polymerase chain reaction; TGF-β1, transforming growth factor β1; and VSMC, vascular smooth muscle cell.
