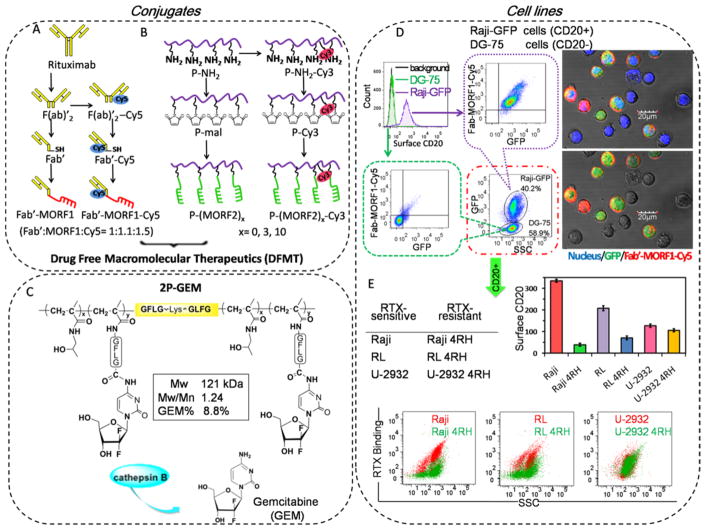
Figure 1.
Conjugates and cell lines. Illustrative structure of (A) Cy5 unlabeled and labeled Fab′-MORF1, (B) Cy3 unlabeled and labeled P-(MORF2)x, and (C) diblock backbone degradable HPMA copolymer-gemcitabine conjugate 2P-GEM. Tetrapeptide GFLG spacer is lysosome enzymatically cleavable. (D) The highly CD20-specific binding of Fab′-MORF1-Cy5 in a co-culture of Raji-GFP cells (expressing green fluorescence) and DG-75 cells. Raji-GFP cells are characterized as CD20 positive cells, and DG-75 cells are characterized as CD20 negative cells. The mixture of Raji-GFP and DG-75 cells were exposed to 1 μM Fab′-MORF1-Cy5 for 1 h. Then Fab′-MORF1-Cy5 binding to different cells (distinguished by GFP expression) were analyzed by flow cytometry and confocal imaging (blue: nuclei; green: GFP; red: Cy5). (E) Surface CD20 expression and RTX binding (at 4 °C) in RTX-sensitive (Raji, RL, U-2932) and RTX-resistant (Raji 4RH, RL 4RH, U-2932 4RH) cell lines. Reductions in CD20 expression and RTX binding were observed in RTX-resistant Raji 4RH and RL 4RH cells, but not U-2932 4RH cells, as compared with their parental RTX-sensitive Raji, RL, and U-2932 cells, respectively.