Figure 4.
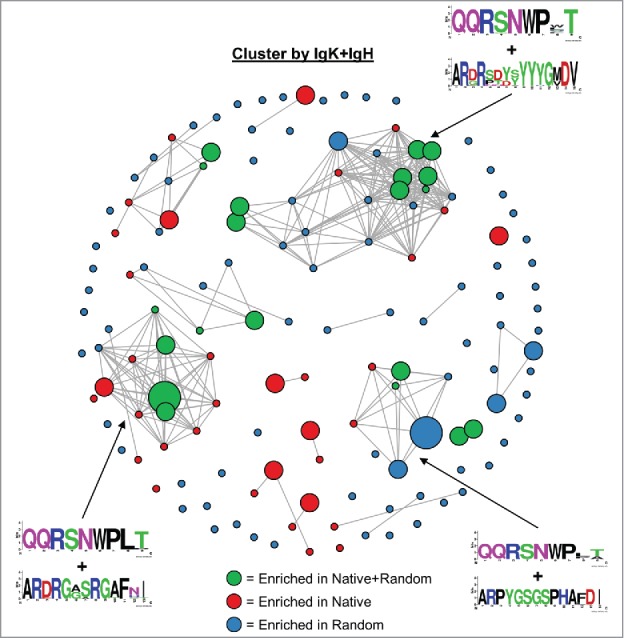
Clonal cluster analysis for FACS-sorted scFv binders. We computed the total number of amino acid differences between each pairwise alignment of FACS-sorted scFv. Edges were drawn only for pairwise alignments with ≤9 amino acid differences. The node for each scFv sequence was sized based on frequency in the FACS-sorted population: small (<1% frequency), medium (1–10% frequency), and large (>10% frequency). scFv isolated from the native+random libraries are indicated with green circles, scFv isolated from the native library only are indicated with red circles, and scFv isolated from the random library only are indicated with blue circles. Web logos of CDR3K + CDR3H sequences of select clusters are shown.
