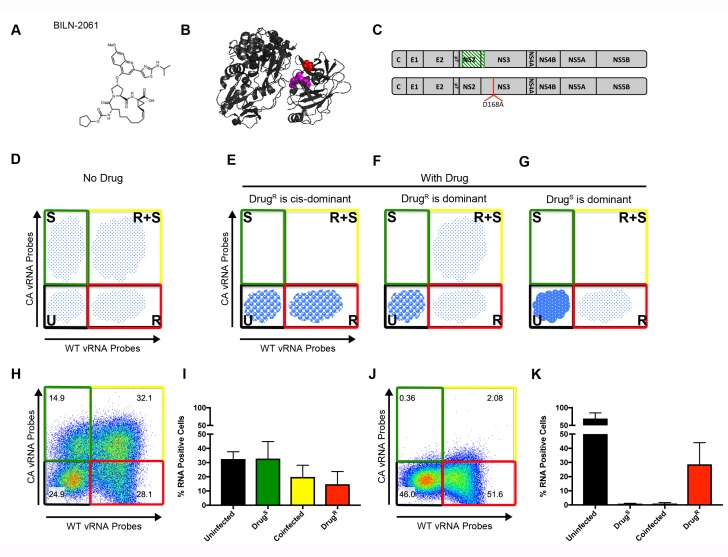
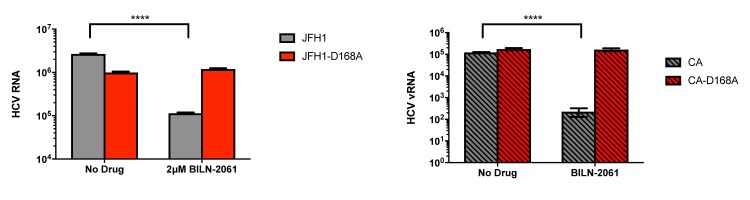
Figure 3. Flow cytometry to test dominance of viruses resistant to NS3 protease inhibitor.
(A) Structure of protease inhibitor BILN-2061. (B) Structure of NS3 protein. D168 (red) is located in the protease domain adjacent to the active site (lavender). D168A confers resistance to BILN-2061. (C) Diagram of CA virus with altered sequence (green hatches) and WT virus with location of D168A mutation identified. (D) The four types of cells present in the absence of inhibitors are uninfected (U), infected with drug-susceptible virus cells (S), infected with both drug-susceptible and drug-resistant virus (S + R) and drug-resistant virus (R). In the presence of a DAA, three outcomes are possible and are indicated by the changing density of the cell populations: (E) Drug-resistance is cis-dominant, (F) Drug resistance is dominant and (G) Drug susceptibility is dominant. Huh7.5.1 cells were coinfected with CA and WT-D168A for 72 hr followed by treatment with (H/I) DMSO or (J/K) 2 μM BILN-2061 for 36 hr. Cells were stained with CA and WT vRNA probes and analyzed by flow cytometry (H, J) and results from three replicates quantified (I, K). NS3 drug-resistance was found to be cis-dominant.