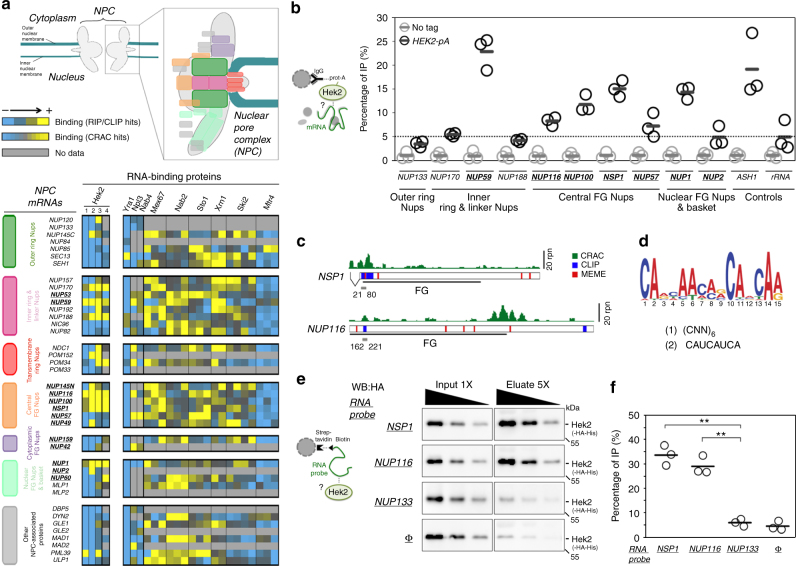
Fig. 1.
The hnRNP K-like protein Hek2 specifically associates with a subset of NPC mRNAs. a Top, Representation of the yeast nuclear pore complex (NPC), showing subcomplexes as colored boxes. Bottom, mRNAs encoding NPC components are sorted by subcomplexes and the strength of their association to the different indicated RNA-binding proteins (RBP) is represented by a color code, as scored in distinct RIP, CLIP or CRAC datasets. Bright yellow indicates the preferred association of a given mRNA to the RBP of interest. For Sto1, Mtr4, Nab2, Mex67, Xrn1 and Ski2, multiple repetitions are displayed27. For Hek2, the results from independent studies are represented: (1)24, (2)26, (3)25, (4)27. FG-Nups appear in bold, underlined. The NUP145 mRNA gives rise to both Nup145-N and Nup145-C nucleoporins and is displayed for each of the according subcomplexes. b Hek2-pA-associated mRNAs were immunopurified and quantified by RT-qPCR using specific primer pairs. Percentages of IP are the ratios between purified and input RNAs, normalized to the amount of purified bait and set to 1 for the “no tag”. Means and individual points (n = 3) are displayed. A schematic representation of the assay is shown. c Overview of Hek2-binding sites on NSP1 and NUP116 mRNAs. The number of CRAC hits (rpn (reads per nucleotide))27, the position of CLIP fragments26 and the occurrences of the binding site found by the MEME analysis are indicated. The positions of the FG-coding region and of minimal Hek2-binding sites used for in vitro pull down (in gray) are represented. The broken line indicates the NSP1 intron. d MEME result from NUP59, NUP116, NUP1, NSP1 and NUP100 sequences. (1, 2): previously identified Hek2-binding sequences24,26. e Left, Schematic representation of the assay. Recombinant HA-tagged Hek2 was incubated with streptavidin beads either naive (Φ) or coated with biotinylated RNA probes encompassing Hek2-binding sites from NSP1 (21–80) or NUP116 (162–221) or a sequence from NUP133 (1429–1488). Right, Decreasing amounts of input and eluate fractions were loaded for quantification. f Percentages of IP are the ratios between Hek2 amounts in the eluate and input fractions, calculated from (e). Means and individual points (n = 3) are displayed. **P < 0.01 (Welch’s t-test)