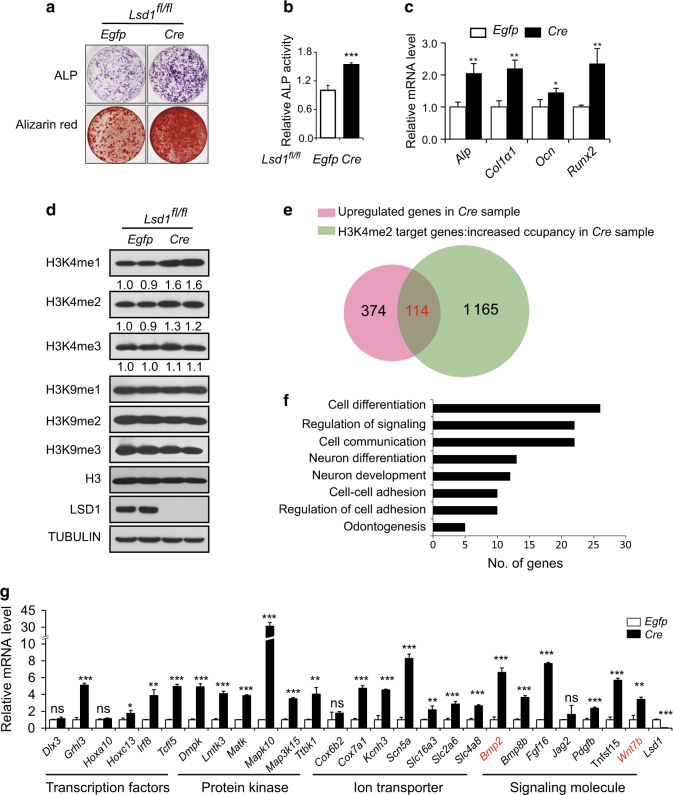
Fig. 4.
Wnt7b and Bmp2 expression is increased in Lsd1 deleted osteoblasts. a–c Lsd1fl/fl calvarial cells derived from P3 mice were infected with Cre-lentivirus or Egfp-lentivirus followed by culturing in osteoblast differentiation media for 14 days. ALP staining (day 7), alizarin red staining (day 14) (a) and quantification of ALP activity (b) of calvarial cells in osteogenic cultures. Data in b are the mean ± SD (n = 6), ***P < 0.001. Unpaired t-test was performed. c mRNA expression of osteoblast marker genes at day7 was analyzed by RT-PCR, n = 3, unpaired t-test, *P < 0.05, **P < 0.01. d Histone H3 lysine4, lysine9 methylation, and LSD1 levels were measured by western blot. e Venn diagram shows the overlap of genes of increased expression level and increased H3K4me2 occupancy at the TSS region in LSD1 mutant osteoblasts. f Gene Ontology (GO) analysis within the 114 genes using the database for annotation, visualization, and integrated discovery (DAVID) bioinformatics tools. The enriched GO biological processes were identified and listed. The enrichment P-value is <0.01. g RT-PCR analysis of the expression levels of candidate genes in RNA-seq samples. Data in g represent the mean ± SD (n = 3) Unpaired t-test was performed. *P < 0.05, **P < 0.01, ***P < 0.001.