Pseudomonas aeruginosa is a dominant and persistent cystic fibrosis pathogen. Although P. aeruginosa is accompanied by other microbes in the airways of cystic fibrosis patients, few cystic fibrosis studies show how P. aeruginosa is affected by the metabolism of other bacteria. Here, we demonstrate that P. aeruginosa generates primary metabolites using substrates produced by another microbe that is prevalent in the airways of cystic fibrosis patients, Rothia mucilaginosa. These results indicate that P. aeruginosa may get a metabolic boost from its microbial neighbor, which might contribute to its pathogenesis in the airways of cystic fibrosis patients.
KEYWORDS: Pseudomonas aeruginosa, Rothia mucilaginosa, metabolite cross-feeding, microbial interactions, polymicrobial infections, stable-isotope-assisted metabolomics
ABSTRACT
Due to a lack of effective immune clearance, the airways of cystic fibrosis patients are colonized by polymicrobial communities. One of the most widespread and destructive opportunistic pathogens is Pseudomonas aeruginosa; however, P. aeruginosa does not colonize the airways alone. Microbes that are common in the oral cavity, such as Rothia mucilaginosa, are also present in cystic fibrosis patient sputum and have metabolic capacities different from those of P. aeruginosa. Here we examine the metabolic interactions of P. aeruginosa and R. mucilaginosa using stable-isotope-assisted metabolomics. Glucose-derived 13C was incorporated into glycolysis metabolites, namely, lactate and acetate, and some amino acids in R. mucilaginosa grown aerobically and anaerobically. The amino acid glutamate was unlabeled in the R. mucilaginosa supernatant but incorporated the 13C label after P. aeruginosa was cross-fed the R. mucilaginosa supernatant in minimal medium and artificial-sputum medium. We provide evidence that P. aeruginosa utilizes R. mucilaginosa-produced metabolites as precursors for generation of primary metabolites, including glutamate.
IMPORTANCE Pseudomonas aeruginosa is a dominant and persistent cystic fibrosis pathogen. Although P. aeruginosa is accompanied by other microbes in the airways of cystic fibrosis patients, few cystic fibrosis studies show how P. aeruginosa is affected by the metabolism of other bacteria. Here, we demonstrate that P. aeruginosa generates primary metabolites using substrates produced by another microbe that is prevalent in the airways of cystic fibrosis patients, Rothia mucilaginosa. These results indicate that P. aeruginosa may get a metabolic boost from its microbial neighbor, which might contribute to its pathogenesis in the airways of cystic fibrosis patients.
OBSERVATION
Cystic fibrosis (CF) patients experience persistent polymicrobial colonization of their airways. Rothia mucilaginosa and Pseudomonas aeruginosa are microbes frequently detected in CF patient airways, and their cooccurrence has been observed in CF patient sputum (1–4). Microbes within polymicrobial infections display complex interactions, such as metabolite cross-feeding (5). For example, P. aeruginosa inefficiently metabolizes host-derived mucins. Rather, P. aeruginosa utilizes mucin degradation products from oral anaerobes to support its growth (6, 7). Still, many studies of CF-associated microbes are conducted under artificial conditions that fail to take into account the nutrient and oxygen gradients found in CF patient airways (8, 9, 25). The lack of overlap between laboratory conditions and CF patient airways is reflected by the differences in growth rates, with estimates of bacterial doubling times being 100-fold times lower in sputum than in standard medium (10). Furthermore, most CF studies focus on single microbes. One primary reason for this is the lack of a robust model to examine the microbial interactions. Stable-isotope-assisted metabolomics analyzes the fate of heavy atoms from stable-isotope-labeled precursors to products, which makes it a suitable approach for monitoring metabolites produced by one microbe when cross-fed to a second microbe. In order to further explore cross-feeding interactions between two CF microbes in a relevant environment, we cross-fed labeled glycolysis products from R. mucilaginosa to P. aeruginosa (8). Both strains were isolated from the sputa of CF patients. We believe that our P. aeruginosa strain is representative of CF strains, as its core genome is similar to that of P. aeruginosa strain PA17 and other CF isolates (11). In an effort to mimic the CF airway environment, R. mucilaginosa was fed labeled glucose in anaerobic and aerobic artificial-sputum media, and the R. mucilaginosa supernatant was fed to P. aeruginosa in nutrient-rich (artificial-sputum medium) under low-nutrient (M9 minimal medium) conditions. As P. aeruginosa lacks some glucose utilization capacities, including a key enzyme involved in glycolysis, phosphofructokinase, we postulated that cross-feeding metabolites from R. mucilaginosa impacts the metabolism of P. aeruginosa (12).
R. mucilaginosa metabolism under aerobic and anaerobic conditions.
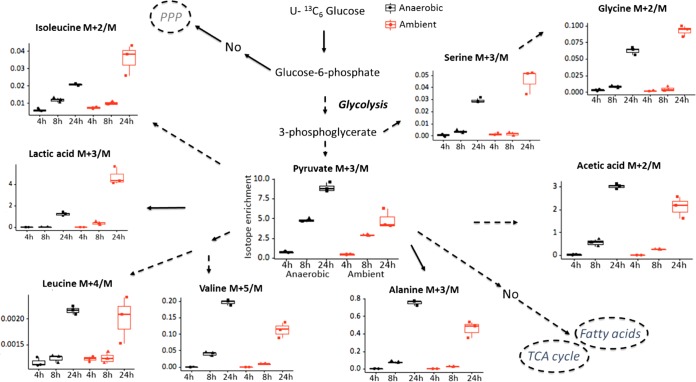
R. mucilaginosa was grown aerobically and anaerobically in artificial-sputum medium (see Text S1 in the supplemental material). Under both anaerobic and aerobic conditions, glucose-derived 13C was incorporated into glycolysis metabolites, namely, lactate and acetate, and some amino acid biosynthesis pathways in R. mucilaginosa (Fig. 1; Table S1). The labeled glucose was not incorporated into the tricarboxylic acid (TCA) cycle, pentose phosphate pathway, or long-chain fatty acid biosynthesis pathways. For most metabolites, 13C incorporation rates were different under different oxygen conditions. For pyruvate, alanine, valine, and acetate, greater label ratios were observed under anaerobic conditions at 24 h. In contrast, lactate, glycine, serine, and isoleucine had greater label ratios under aerobic conditions at 24 h. The incorporation of [U-13C6]glucose into leucine biosynthesis was not impacted by oxygen conditions. Carbon fate in R. mucilaginosa diverged after 3-phosphoglycerate. The 13C label was incorporated into serine and glycine, or into pyruvate, the precursor for lactate, acetate, and some amino acids.
FIG 1 .
Glucose-derived 13C was incorporated into pyruvate, lactate, acetate, alanine, valine, serine, glycine, leucine, and isoleucine in R. mucilaginosa under both anaerobic and ambient-oxygen conditions. M+2, M+3, M+4, and M+5 indicate compounds that contained 2, 3, 4, and 5 13C atoms, respectively. Isotope enrichment means an abundance of labeled ion/unlabeled ion (corrected for natural abundance). Isotope enrichment was greater at 24 h than at 8 h or 4 h. For pyruvate, alanine, valine, and acetate, greater isotope enrichment was observed under anaerobic conditions at 24 h. For lactate, glycine, serine, and isoleucine, greater isotope enrichment was observed under ambient-oxygen conditions at 24 h. The incorporation of glucose-derived 13C into leucine biosynthesis was not affected by oxygen conditions. Dashed lines and solid lines indicate multiple steps and one metabolic step(s) needed to obtain the metabolite, respectively. Error bars, means ± standard deviations (SD) (n = 3 bacterial cultures per group); TCA, citric acid cycle; PPP, pentose phosphate pathway.
Supplemental experimental details. Download TEXT S1, PDF file, 0.2 MB (177KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isotope enrichment of metabolites in R. mucilaginosa under anaerobic and aerobic oxygen condition. Download TABLE S1, XLSX file, 0.04 MB (47.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cross-feeding interactions between R. mucilaginosa and P. aeruginosa.
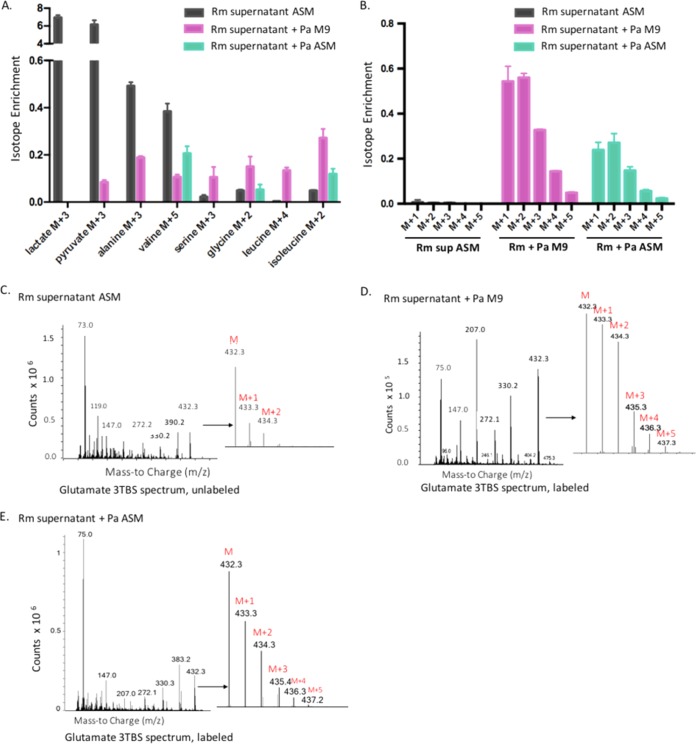
In order to study the impact of R. mucilaginosa metabolites on P. aeruginosa, we cross-fed supernatant from an aerobic 48-h R. mucilaginosa culture to P. aeruginosa grown under low-nutrient conditions (M9 minimal medium) and nutrient-rich conditions (artificial-sputum medium). P. aeruginosa was grown for 120 h before the cells were harvested in order to recapitulate the low growth rates of bacteria in CF patient sputa (10). The R. mucilaginosa supernatant included labeled lactate, pyruvate, and alanine (Fig. 2A; Fig. S1; Table S2). P. aeruginosa utilized R. mucilaginosa-derived metabolites to produce metabolites in M9 minimal medium and artificial-sputum medium. For example, although labeled lactate was found in the R. mucilaginosa supernatant, it was not detected in P. aeruginosa cultures, suggesting that P. aeruginosa consumed R. mucilaginosa-derived lactate (Fig. 2A; Fig. S1; Table S2). P. aeruginosa utilization of lactate and other fermentation products has been observed in other studies (6, 13). Since lactate levels have been reported as an indicator of CF patient response to antibiotic therapy, the finding that P. aeruginosa consumes lactate derived from another CF microbe may have clinical implications (14).
FIG 2 .
Cross-feeding interactions between R. mucilaginosa and P. aeruginosa. M+1, M+2, M+3, M+4, and M+5 indicate compounds that contained 1, 2, 3, 4, and 5 13C atoms, respectively. Error bars, means ± SD (n = 3 bacterial cultures per group). (A) Labeled lactate was found in the R. mucilaginosa (Rm) supernatant but not in P. aeruginosa (Pa) cells. In M9 minimal medium, P. aeruginosa cells contained isotopically enriched pyruvate, alanine, valine, serine, glycine, leucine, and isoleucine. In artificial-sputum medium, P. aeruginosa cells contained isotopically enriched valine, glycine, and isoleucine. (B) Although the R. mucilaginosa supernatant contained only unlabeled glutamate, labeled glutamate was detected in the P. aeruginosa cells grown in artificial-sputum medium and M9 minimal medium. (C to E) Glutamate spectrum for the R. mucilaginosa supernatant (C), P. aeruginosa grown in M9 minimal medium spiked with the R. mucilaginosa supernatant (D), and P. aeruginosa grown in artificial-sputum medium spiked with the R. mucilaginosa supernatant (E).
Abundances of isotope-enriched metabolites for R. mucilaginosa grown in artificial-sputum medium (Rm), P. aeruginosa grown in M9 minimal medium spiked with the R. mucilaginosa supernatant (Rm_Pa_M9), and P. aeruginosa grown in artificial-sputum medium spiked with the R. mucilaginosa supernatant (Rm_Pa_ASM). Download FIG S1, PDF file, 0.2 MB (215.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isotope enrichment of metabolites in the R. mucilaginosa supernatant and P. aeruginosa fed the R. mucilaginosa supernatant. Download TABLE S2, XLSX file, 0.04 MB (46.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Labeled metabolites detected in P. aeruginosa cells grown in minimal medium included pyruvate, alanine, valine, serine, glycine, leucine, and isoleucine (Fig. 2A; Fig. S1; Table S2). In addition, isotope enrichment for serine, glycine, leucine, and isoleucine was greater in P. aeruginosa cells than in the supernatant of R. mucilaginosa, indicating that P. aeruginosa biosynthesized those metabolites. In contrast, when P. aeruginosa was grown in artificial-sputum medium, P. aeruginosa had higher levels of a single isotope-enriched amino acid (isoleucine) than occurred in the R. mucilaginosa supernatant (Fig. 2A; Fig. S1; Table S2). Interestingly, although the R. mucilaginosa supernatant contained only unlabeled glutamate (Fig. 2B and C; Fig. S2; Table S3), labeled glutamate was detected in both P. aeruginosa cultures (Fig. 2B, D, and E; Fig. S2; Table S3). This suggests that P. aeruginosa biosynthesized glutamate from 13C sources in the R. mucilaginosa supernatant even in a nutrient-rich background with initially freely available glutamate (Text S1).
Abundances of isotope-enriched glutamate ions for R. mucilaginosa grown in artificial-sputum medium (Rm), P. aeruginosa grown in M9 minimal medium spiked with the R. mucilaginosa supernatant (Rm_Pa_M9), and P. aeruginosa grown in artificial-sputum medium spiked with the R. mucilaginosa supernatant (Rm_Pa_ASM). Download FIG S2, PDF file, 0.2 MB (190.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isotope enrichment of glutamate in the R. mucilaginosa supernatant and P. aeruginosa fed the R. mucilaginosa supernatant. Download TABLE S3, XLSX file, 0.04 MB (40.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Glutamate provides a link between nitrogen and carbon metabolism by serving as a major amine group donor in transamination reactions for the synthesis of additional amino acids and nucleosides. In Escherichia coli, up to 88% of the total nitrogen that ends up in a biomass comes from glutamate, and the cellular glutamate pool needs to be kept high to drive the transamination reactions (15). In P. aeruginosa specifically, glutamate is a component of the cell wall and may play a role in P. aeruginosa virulence (16). Glutamate enhanced the yield of a virulence factor, exotoxin A (17), and induced swarming motility in P. aeruginosa on semisolid surfaces (18). More recently, glutamate-induced dispersion via c-di-GMP signaling pathways has been suggested (19). Glutamate might be derived from glutamine or alpha-ketoglutarate (20–22). However, the abundance of these two compounds was below the limit of quantification in this study. Future studies are needed to examine the biosynthesis pathways of glutamate and its role in the metabolism and physiology of P. aeruginosa. In summary, this study provides evidence that metabolite cross-feeding exists between R. mucilaginosa and P. aeruginosa, two common microorganisms found in polymicrobial communities in CF patient airways. The results from our study provide evidence that the physiology of CF pathogens can be influenced by the metabolic capabilities of other nearby microorganisms, even in a nutrient-rich environment, which can be tracked with stable-isotope-labeled metabolomics.
Culture conditions and metabolomics.
The bacterial strains chosen for this study were isolated from CF patients at the UCSD Adult CF Clinic: Pseudomonas aeruginosa PaFLR01 and Rothia mucilaginosa RmFLR01 (11, 23). First, we took time points from R. mucilaginosa cultures to examine the kinetics of metabolites in glycolysis, the TCA cycle, amino acid biosynthesis, short- and long-chain fatty acid biosynthesis, and the pentose phosphate pathway in R. mucilaginosa, which was grown in triplicate in artificial-sputum medium (24) spiked with 100 mM [U-13C6]d-glucose (Sigma-Aldrich and Cambridge Isotope Laboratory) under anaerobic and aerobic oxygen conditions (5% CO2) at 37°C. R. mucilaginosa cells were harvested at 4 h, 8 h, and 24 h. For the metabolite cross-feeding study, R. mucilaginosa was grown in the same medium aerobically for 48 h. The R. mucilaginosa supernatant was collected by filtering the culture, and the supernatant was diluted 10-fold in M9 minimal medium supplemented with succinate and in fresh artificial-sputum medium. P. aeruginosa was grown in triplicate aerobically, and the cells were harvested at 120 h. Metabolite extraction and data acquisition were carried out by following West Coast Metabolomics Center standard operating procedures (Text S1). Agilent MassHunter quantitative analysis software (v. B.07.00) was used for raw data processing. Natural abundance was corrected when isotope enrichment was calculated.
ACKNOWLEDGMENTS
This work was supported as a pilot project by the NIH (grant DK097154). K.L.W. is supported by a Gilead CF Research Scholars Award (app_00b072). T.G. is supported through the National Science Foundation’s Integrative Graduate Education and Research Traineeship (IGERT) program (grant DGE-1144901).
REFERENCES
- 1.Blainey PC, Milla CE, Cornfield DN, Quake SR. 2012. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci Transl Med 4:153ra130. doi: 10.1126/scitranslmed.3004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim YW, Schmieder R, Haynes M, Furlan M, Matthews TD, Whiteson K, Poole SJ, Hayes CS, Low DA, Maughan H, Edwards R, Conrad D, Rohwer F. 2013. Mechanistic model of Rothia mucilaginosa adaptation toward persistence in the CF lung, based on a genome reconstructed from metagenomic data. PLoS One 8:e64285. doi: 10.1371/journal.pone.0064285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, Zhang Y, Surendra A, Gong Y, Elizabeth Tullis D, Yau YC, Waters VJ, Hwang DM, Guttman DS. 2015. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 5:10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey MM, Rumbaugh KP, Whiteley M. 2011. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog 7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn JM, Niccum D, Dunitz JM, Hunter RC. 2016. Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog 12:e1005846. doi: 10.1371/journal.ppat.1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn JM, Phan C, Hunter RC. 2017. Genome-wide survey of Pseudomonas aeruginosa PA14 reveals a role for the glyoxylate pathway and extracellular proteases in the utilization of mucin. Infect Immun 85:e00182-17. doi: 10.1128/IAI.00182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garg N, Wang M, Hyde E, da Silva RR, Melnik AV, Protsyuk I, Bouslimani A, Lim YW, Wong R, Humphrey G, Ackermann G, Spivey T, Brouha SS, Bandeira N, Lin GY, Rohwer F, Conrad DJ, Alexandrov T, Knight R, Dorrestein PC. 2017. Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22:705–716.e4. doi: 10.1016/j.chom.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. 2015. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen-sulfide formation. mBio 6:e00767-15. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopf SH, Sessions AL, Cowley ES, Reyes C, Van Sambeek LV, Hu Y, Orphan VJ, Kato R, Newman DK. 2016. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A 113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan J, Gallagher T, Oliver A, England W, Whiteson K. 29 March 2018. Fermentation products in the cystic fibrosis airways induce aggregation and dormancy-associated expression profiles in a CF clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol Lett doi: 10.1093/femsle/fny082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C. 2015. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkataraman A, Rosenbaum MA, Perkins SD, Werner JJ, Angenent LT. 2011. Metabolite-based mutualism between Pseudomonas aeruginosa PA14 and Enterobacter aerogenes enhances current generation in bioelectrochemical systems. Energy Environ Sci 4:4550–4559. doi: 10.1039/c1ee01377g. [DOI] [Google Scholar]
- 14.Bensel T, Stotz M, Borneff-Lipp M, Wollschläger B, Wienke A, Taccetti G, Campana S, Meyer KC, Jensen PØ, Lechner U, Ulrich M, Döring G, Worlitzsch D. 2011. Lactate in cystic fibrosis sputum. J Cyst Fibros 10:37–44. doi: 10.1016/j.jcf.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke K, Gray GW, Reaveley DA. 1967. The cell walls of Pseudomonas aeruginosa. General composition. Biochem J 105:749–754. doi: 10.1042/bj1050749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somerville G, Mikoryak CA, Reitzer L. 1999. Physiological characterization of Pseudomonas aeruginosa during exotoxin A synthesis: glutamate, iron limitation, and aconitase activity. J Bacteriol 181:1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köhler T, Curty LK, Barja F, van Delden C, Pechère JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu Roy A, Sauer K. 2014. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol 94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helling RB. 1998. Pathway choice in glutamate synthesis in Escherichia coli. J Bacteriol 180:4571–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan D. 2007. Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci U S A 104:9475–9480. doi: 10.1073/pnas.0703360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunka K, Commichau FM. 2012. Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol Microbiol 85:213–224. doi: 10.1111/j.1365-2958.2012.08105.x. [DOI] [PubMed] [Google Scholar]
- 23.Phan J, Meinardi S, Barletta B, Blake DR, Whiteson K. 2017. Stable isotope profiles reveal active production of VOCs from human-associated microbes. J Breath Res 11:017101. doi: 10.1088/1752-7163/aa5833. [DOI] [PubMed] [Google Scholar]
- 24.Quinn RA, Whiteson K, Lim YW, Salamon P, Bailey B, Mienardi S, Sanchez SE, Blake D, Conrad D, Rohwer F. 2015. A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J 9:1024–1038. doi: 10.1038/ismej.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental experimental details. Download TEXT S1, PDF file, 0.2 MB (177KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isotope enrichment of metabolites in R. mucilaginosa under anaerobic and aerobic oxygen condition. Download TABLE S1, XLSX file, 0.04 MB (47.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Abundances of isotope-enriched metabolites for R. mucilaginosa grown in artificial-sputum medium (Rm), P. aeruginosa grown in M9 minimal medium spiked with the R. mucilaginosa supernatant (Rm_Pa_M9), and P. aeruginosa grown in artificial-sputum medium spiked with the R. mucilaginosa supernatant (Rm_Pa_ASM). Download FIG S1, PDF file, 0.2 MB (215.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isotope enrichment of metabolites in the R. mucilaginosa supernatant and P. aeruginosa fed the R. mucilaginosa supernatant. Download TABLE S2, XLSX file, 0.04 MB (46.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Abundances of isotope-enriched glutamate ions for R. mucilaginosa grown in artificial-sputum medium (Rm), P. aeruginosa grown in M9 minimal medium spiked with the R. mucilaginosa supernatant (Rm_Pa_M9), and P. aeruginosa grown in artificial-sputum medium spiked with the R. mucilaginosa supernatant (Rm_Pa_ASM). Download FIG S2, PDF file, 0.2 MB (190.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isotope enrichment of glutamate in the R. mucilaginosa supernatant and P. aeruginosa fed the R. mucilaginosa supernatant. Download TABLE S3, XLSX file, 0.04 MB (40.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.