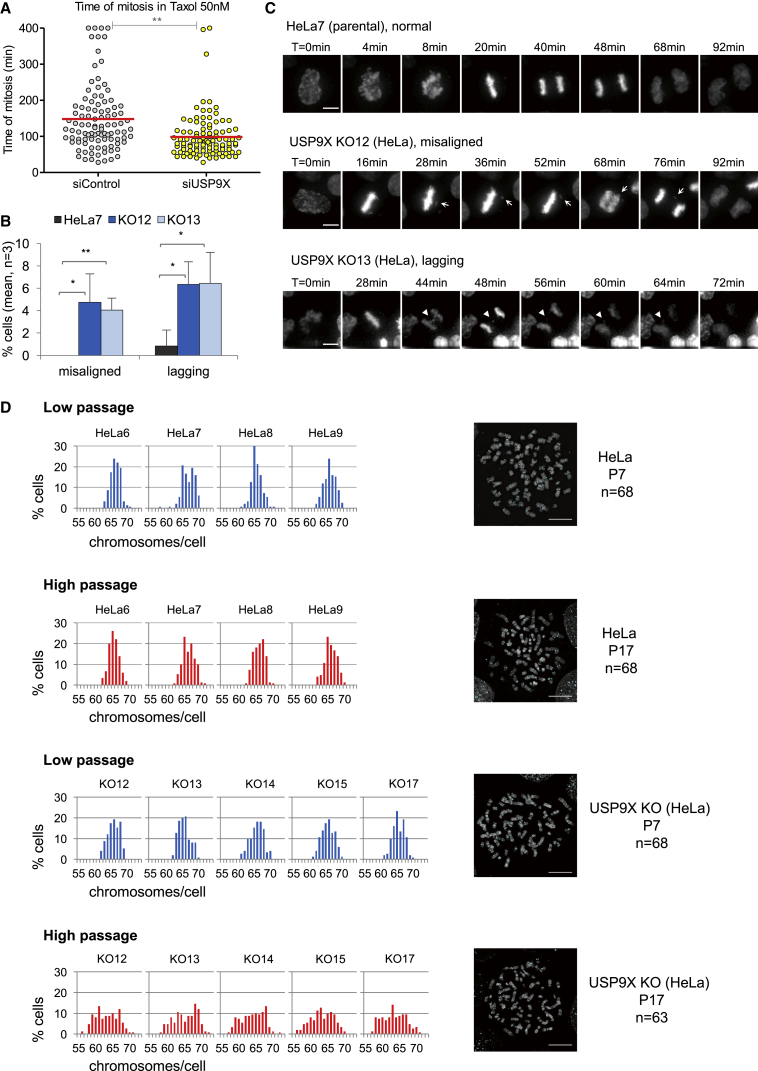
Figure 5.
Loss of USP9X Induces Segregation Defects and Chromosomal Instability
(A) Average mitotic duration in Taxol-arrested (50 nM) U2OS cells treated with control or USP9X siRNAs. The graph shows the mean values (±SD) from two independent experiments with 50 cells quantified for each condition. ∗∗p < 0.01, unpaired Student’s t test.
(B and C) Quantification (B) and representative images (C) of misaligned (arrows) and lagging (arrowheads) chromosomes in HeLa parental cells (HeLa7) and USP9X knock out clones (KO12, KO13). The graph shows the mean values (±SD) from three independent experiments. At least 230 cells were analyzed per cell line with >60 cells per experiment. DNA was visualized using SiR-DNA, and cells were imaged every 4 min. ∗p < 0.05 and ∗∗p < 0.01, unpaired Student’s t test. Scale bar, 10 μM.
(D) Chromosome counts from metaphase spreads in parental (HeLa 6, 7, 8, 9) or USP9X knockout (KO12, KO13, KO14, KO15, KO17) HeLa clones at early (P7) and late (P17) passage. The combined data for 150 cells from three independent experiments are shown; >40 cells were analyzed per experiment. Representative images of metaphase spreads from parental and USP9X knockout cells at early and late passage are shown (right). Scale bar, 10 μM.
See also Figure S5.