Figure 3.
In Vitro Immortalization Assay
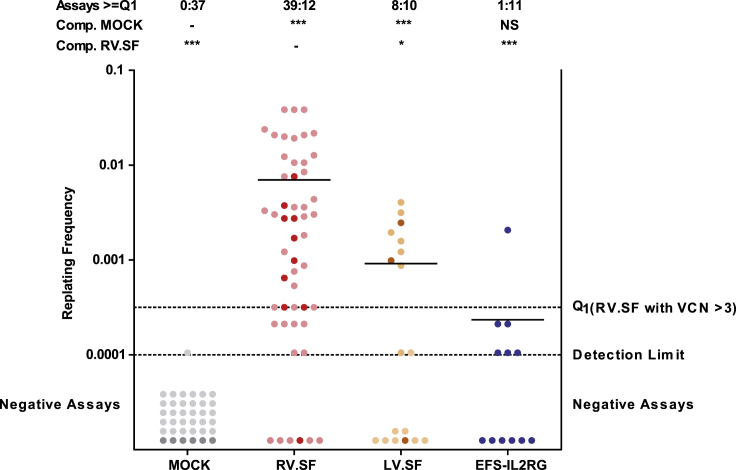
Insertional mutants are identified by clonal outgrowth on a replating assay, where non-immortalized cells do not grow (negative, below detection limit). The number of positive wells is used to calculate the replating frequency (RF) according to Poisson statistics. Positive assays above the Q1 level (replating frequency [RF] of 3.17 × 10−4) are counted as positive. Each dot represents one assay. Control data (MOCK, RV.SF, and LV.SF) from previous experiments conducted under the same standard operation procedure are included. Darker colors mark the actual assays and lighter colors indicate the meta data. Bars show the mean RF. Above the graph, the ratio of assays above and below the Q1 level are given together with a statistical analysis on the incidence of positive and negative plates. EFS-IL2RG had a significantly lower mutagenic potential compared to RV.SF and was indistinguishable from MOCK. (NS, not significant; ***p < 0.001 and *p < 0.05, Fisher’s exact test with Benjamini-Hochberg multiple comparison correction).