Summary
The transition from naive to primed state of pluripotent stem cells is hallmarked by epithelial-mesenchymal transition, metabolic switch from oxidative phosphorylation to aerobic glycolysis, and changes in the epigenetic landscape. Since these changes are also seen as putative hallmarks of neoplastic cell transformation, we hypothesized that oncogenic pathways may be involved in this process. We report that the activity of RAS is repressed in the naive state of mouse embryonic stem cells (ESCs) and that all three RAS isoforms are significantly activated upon early differentiation induced by LIF withdrawal, embryoid body formation, or transition to the primed state. Forced expression of active RAS and RAS inhibition have shown that RAS regulates glycolysis, CADHERIN expression, and the expression of repressive epigenetic marks in pluripotent stem cells. Altogether, this study indicates that RAS is located at a key junction of early ESC differentiation controlling key processes in priming of naive cells.
Keywords: RAS, pluripotent stem cells, naive, primed, metabolism, cancer
Graphical Abstract
Highlights
-
•
RAS is activated upon early differentiation of mouse embryonic stem cells (mESCs)
-
•
RAS repression by LIF prevents early differentiation
-
•
RAS regulates the transition from naive to primed state of mESCs
-
•
RAS regulates glycolysis, chromatin remodeling, and CADHERIN expression
Altshuler et al. report that RAS activation positively regulated key processes of naive-primed transition of mouse embryonic stem cells, including changes in metabolism, chromatin remodeling, and the switch in CADHERIN expression. Pharmacological inhibition of RAS attenuated cellular priming, suggesting that RAS inhibition may be potentially useful for converting human cells into ground state and for efficient somatic cellular reprogramming.
Introduction
Pluripotency, defined as the ability of a cell to generate all cell types in the adult organism, is a transient feature of early embryonic development. It has been shown that pluripotency may exist in at least two fundamentally distinct states, namely, “naive” and “primed” (Gafni et al., 2013, Nichols and Smith, 2009, Takashima et al., 2015, Theunissen et al., 2014). Although both naive and primed pluripotent stem cells (PSCs) are able to form the three germ layers in vitro and in a teratoma assay, only naive PSCs are able to efficiently contribute to the formation of chimeric animals (Rossant, 2008). Naive state culture of murine embryonic stem cells (mESCs) can be sustained in the presence of serum and leukemia inhibitory factor (fetal calf serum [FCS]/LIF). However, a more uniform “ground state” culture that mirrors better the undifferentiated transcriptional and epigenetic landscape of pre-implantation epiblast cells can be achieved in the presence of a combination of LIF and the inhibitors of MEK and GSKβ (2i/LIF) (Hackett and Azim Surani, 2014, Nichols and Smith, 2009, Weinberger et al., 2016, Wray et al., 2010, Ying et al., 2008).
In contrast to mouse PSCs (mPSCs) that display features of naive state, human PSCs (hPSCs) are believed to be stabilized in a primed state of pluripotency. Cells that are at naive state are considered to be more amenable for genetic manipulation, and are able to differentiate more uniformly. Thus, many efforts have been made to characterize the molecular pathways regulating pluripotency states (Boroviak et al., 2014, Buecker et al., 2014, Guo et al., 2009, Hackett and Azim Surani, 2014, Kalkan and Smith, 2014, Weinberger et al., 2016), and in particular to convert primed hPSCs into naive state (Chan et al., 2013, Gafni et al., 2013, Takashima et al., 2015, Theunissen et al., 2014, Ware et al., 2014, Yang et al., 2017). Yet, there is a controversy regarding the quality of the resulting cells, to what extent they appropriately reflect preimplantation cells, while culture conditions typically require the combination of multiple soluble factors and inhibitors. Therefore, a better understanding of the signaling pathways that control self-renewal at the different states of pluripotency is necessary. Optimized culture of naive cells would allow an appropriate study of early development and lineage commitments using PSCs and their efficient application.
Interestingly, the transition from naive to primed state is accompanied by cellular changes that are to some extent similar to cancer cell transformation. These changes include metabolic switch from oxidative phosphorylation to anaerobic glycolysis, marks of epithelial-mesenchymal transition (EMT), and drastic epigenetic changes, suggesting that this process may be mediated by oncogenic pathways. The role of RAS proteins has been extensively studied in the field of cancer cell biology; however, their involvement in stem cells and cellular reprogramming remained largely unexplored. The three RAS isoforms, namely, H-RAS, K-RAS, and N-RAS, are encoded by three separate genes and they possess many overlapping roles, although some isoform-specific features has been reported (Prior and Hancock, 2012, Schubbert et al., 2007). RAS proteins act as molecular switches, alternating between inactive guanosine diphosphate (GDP)-bound state and active guanosine triphosphate (GTP)-bound state. Upon receptor-mediated signal transduction, RAS proteins become active (GTP bound) and undergo allosteric change in their conformation, allowing them to recruit a large set of proteins known as Ras effector proteins (Mitin et al., 2005, Vigil et al., 2010). Among these are mitogen-activated protein kinase and phosphatidylinositol 3-kinase (PI3K), which regulate a cascade of signals leading to a wide range of cellular responses, including growth, differentiation, inflammation, survival, and apoptosis. Although RAS proteins are involved in many biological processes in health and disease, their involvement in early embryogenesis and ESC differentiation remained largely unexplored.
Here, we show that all three RAS isoforms are activated upon early ESC differentiation. While low RAS activity hallmarks the naive state of pluripotency, RAS activation is necessary and sufficient to induce key features of differentiation, indicating that RAS is located at a key junction of this process. Inhibition of RAS significantly attenuates differentiation, while its ectopic expression is sufficient to induce differentiation, suggesting that RAS plays a role at early embryogenesis and that it may serve as a key target for cellular reprogramming into the naive state.
Results
RAS Regulates Early Differentiation of mESCs
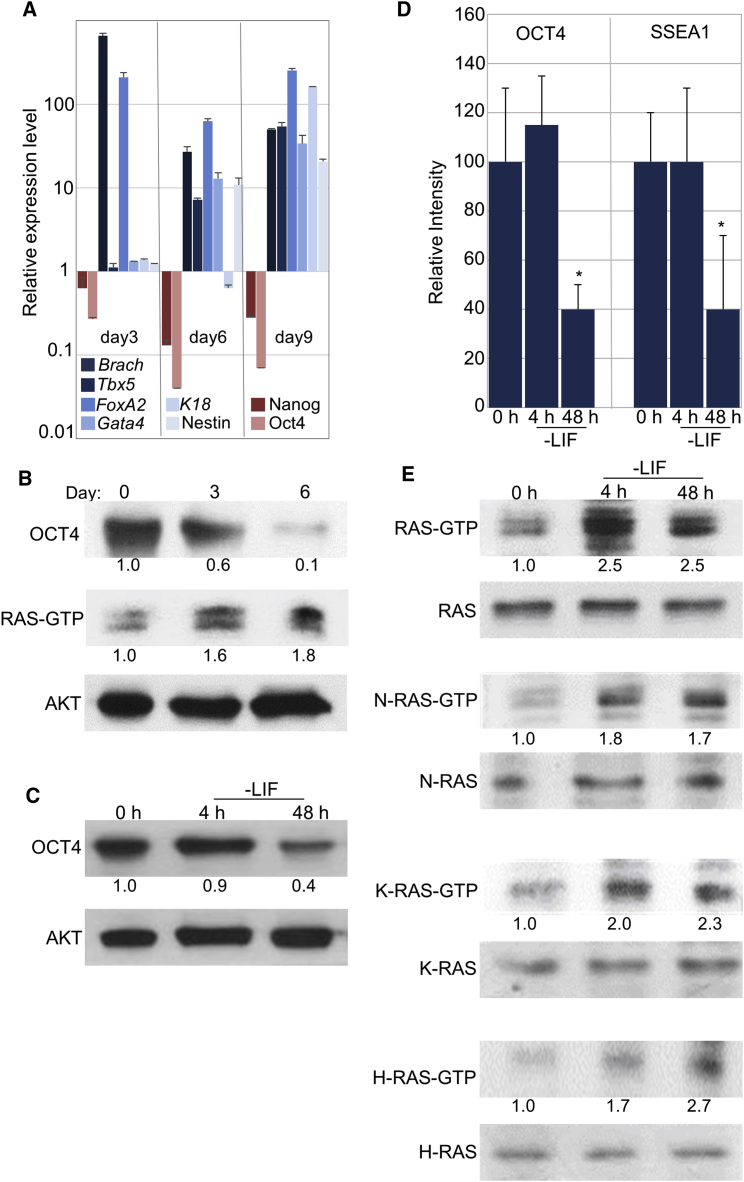
To examine the expression pattern and activity of RAS in pluripotency and early differentiation, we used mESCs (CGR8 cells) that were grown in self-renewal conditions in the presence of serum and LIF (FCS/LIF). Cells were differentiated into the three embryonic germ layers through embryonic body (EB) formation for up to 9 days, as detailed in the Experimental Procedures. As expected, differentiation was accompanied by downregulation of pluripotency markers (OCT4 and NANOG), and increased expression of markers of the three germ layers (Figure 1A). To examine the levels of active (GTP bound) RAS proteins, cell lysates were prepared for RAS-GTP pull-down assay followed by western blot analysis using pan-RAS antibody, as reported previously (Shalom-Feuerstein et al., 2005, Shalom-Feuerstein et al., 2008). RAS activity significantly increased upon differentiation concomitantly with a decrease in OCT4 (Figure 1B), suggesting that RAS may be involved in an early exit from the undifferentiated state. To further investigate the link between RAS activity and self-renewal of mESCs, cells grown in FCS/LIF were deprived of LIF for 4 or 48 hr. No change in the expression of pluripotency markers OCT4 and SSEA1 was evident 4 hr following LIF removal. However, a significant reduction in these proteins was evident by 48 hr (Figures 1C, 1D, and S1). To further explore the activity of each of the three canonical RAS isoforms, we used isoform-specific antibodies. All three RAS isoforms were activated as early as 4 hr following LIF removal, while the total protein levels of each isoform remained unaffected (Figure 1E). Given that the activation of RAS proteins preceded the reduction in the expression of pluripotency markers, these data suggest that RAS may be involved in the control of early differentiation.
Figure 1.
RAS Activity Is Induced in Early Differentiation
(A) Real-time PCR analysis showing the relative expression of markers of pluripotency (Nanog and Oct4) and markers of the three germ layers (Brach, Tbx5, FoxA2, Gata4, K18 and Nestin) at the indicated days of embryoid body (EB) differentiation, compared with undifferentiated cells.
(B) Differentiated cells (EBs) were lysed at the indicated days. RAS-GTP pull-down assay followed by western blot analysis of active pan-RAS (RAS-GTP) or western blot analysis of OCT4 or AKT (protein loading) is shown.
(C) Western blot analysis of OCT4 or AKT (protein loading) at the indicated time points following LIF removal.
(D) Quantification of expression of OCT4 and SSEA1 proteins (data shown in Figure S1) at the indicated time points following LIF removal. Quantification was performed using a Nikon NIS-Element D, as detailed in the Experimental Procedures.
(E) RAS-GTP pull-down assay was followed by western blot analysis using pan-RAS antibody (RAS-GTP) or using the indicated RAS isoform-specific antibodies (H-RAS-GTP, K-RAS-GTP, or N-RAS-GTP). The total expression of pan-RAS (RAS) or of each isoform (H-RAS, K-RAS, or N-RAS) was examined by western blot.
(B, C, and E) Values represent densitometry analysis of at least three independent experiments. Data shown are mean ± SD from three independent experiments. ∗p < 0.05 statistically significant by Student's t test.
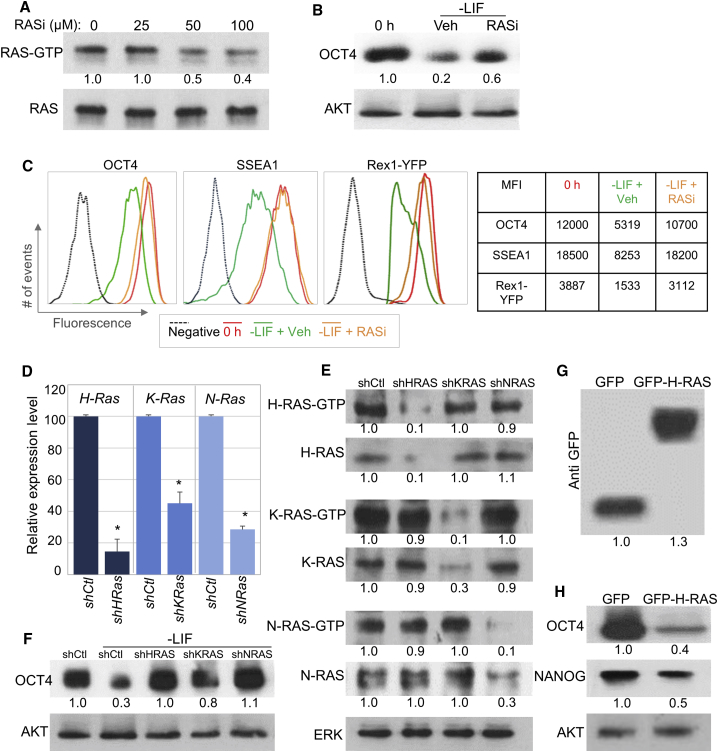
To test this hypothesis further, we examined the effect of RAS inhibition using a pharmacological inhibitor targeting all RAS isoforms. The RAS inhibitor (RASi) farnesylthiosalicyclic acid disrupts RAS membrane anchorage and inhibits tumor cell growth (Bustinza-Linares et al., 2010, Cox et al., 2014, Shalom-Feuerstein et al., 2004, Weisz et al., 1999). In line with previous reports, doses of 50–100 μM significantly reduced the levels of RAS-GTP in mESCs (Figure 2A). To explore the role of RAS in mESC differentiation, cells that were differentiated in the absence of LIF were treated with RASi (75 μM) or vehicle (Veh) as control. RASi significantly attenuated OCT4 reduction following LIF withdrawal (Figure 2B). To corroborate these data, cells that were differentiated in the absence of LIF and in the presence of RASi or Veh were immunnostained for SSEA1 or OCT4 and then subjected to flow cytometry analysis. Once again, RASi prevented the differentiation-induced decrease in OCT4 (Figure 2C, left) and SSEA1 (Figure 2C, middle). Likewise, similar results were obtained using mESCs that stably express Rex1-YFP reporter (Figure 2C, right); thus, the significant decrease in YFP signal upon differentiation was attenuated in the presence RASi (Figure 2C). To validate our data and further explore the link between RAS and cell differentiation, we examined the impact of knock down or overexpression of RAS. Efficient knock down of each of the three RAS isoforms was established using lentiviral infections (Figures 2D, 2E, S2A, and S2B). Cells that were infected with control lentiviral vector showed a marked reduction in OCT4 protein following LIF starvation (Figure 2F). However, the decline in OCT4 levels was significantly attenuated following infection with the small hairpin RNA (shRNA) against RAS-specific isoforms (Figure 2F). These data suggest that the induction of RAS-GTP upon LIF removal is required for ESC differentiation. In other words, in the presence of LIF, RAS is repressed and the low activity of RAS may be necessary for preventing an exit from pluripotency. To further test this hypothesis we examined whether RAS expression alone is sufficient to induce differentiation. mESCs were transfected with a plasmid encoding for GFP fused to H-RAS (GFP-H-RAS) or GFP alone as control. Cells were harvested 48–72 hr later, GFP-positive cells were sorted by flow cytometry (Figure S3), and the expression of GFP-RAS or GFP was validated by western blot analysis (Figure 2G). Notably, both OCT4 and NANOG were significantly reduced following transfection of GFP-RAS (Figure 2H). Altogether, these data strongly suggest that RAS induces early events of mESC differentiation.
Figure 2.
RAS Repression by LIF Prevents Early Differentiation
(A–C) mESCs were differentiated in the absence of LIF for 72 hr and in the presence of the RAS inhibitor farnesylthiosalicyclic acid (RASi, at the indicated concentrations or 75 μM if not mentioned) or vehicle (Veh) as control. (A) The relative RAS-GTP or total RAS protein levels were analyzed in cells treated with the indicated concentration of RASi. (B) Western blots of OCT4 and AKT (loading control). (C) mESCs or Rex1-YFP-expressing mESCs were differentiated in the absence of LIF and in the presence of RASi (75 μM) or Veh for 72 hr. Cells were stained for SSEA1 and OCT4, and YFP fluorescence was measured by flow cytometry. The table shows mean fluorescence of each staining or YFP.
(D–F) mESCs were infected with shRNAs against the indicated RAS isoform or control shRNA and the levels of the relevant RAS isoforms were examined by real-time PCR analysis (D) and by western blot (E). Western blot of OCT4 and AKT of infected mESCs differentiated in the absence of LIF for 72 hr (F).
(G and H) mESCs were transfected with a plasmid encoding for GFP fused to H-RAS (GFP-H-RAS) or GFP as control. GFP-positive cells were sorted by flow cytometry, lysed, and immunoblotted using anti-GFP antibody (G) and pluripotency markers (H). AKT or ERK were used as a loading control.
(A, B, and E–H) Values represent densitometry analysis of at least three independent experiments. Data shown are mean ± SD from three independent experiments. ∗p < 0.05 statistically significant by Student's t test.
RAS Modulates the Transition from Naive to Primed State
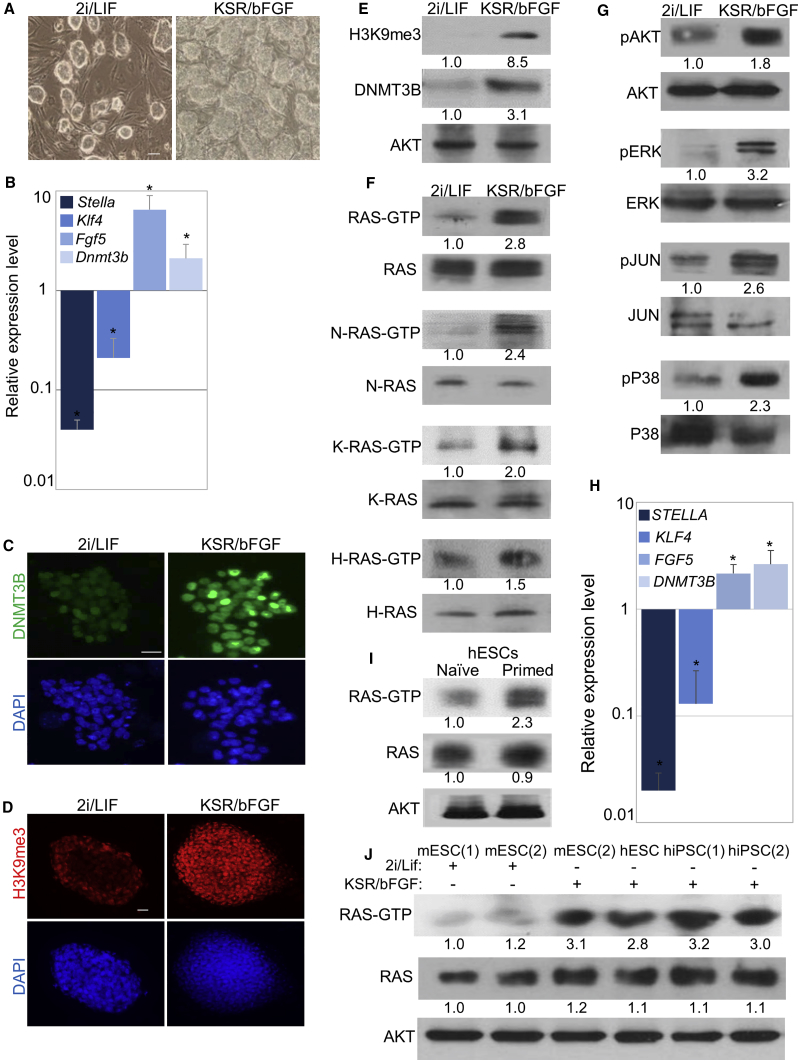
To further test the role of RAS in early events of mESC differentiation, we checked the possibility that RAS may be involved in the transition from the naive ground state to the primed state of pluripotency. To address this possibility, mESCs that were grown in naive (ground state) conditions (2i/LIF) were switched into primed state by cultivating the cells for 10 passages in the presence of knockout serum and basic fibroblast growth factor (KSR/bFGF), as reported previously (Hanna et al., 2009). As expected, the transition to the primed state was accompanied by morphological changes (Figure 3A). Real-time qPCR analysis confirmed the decrease in expression of markers of the naive state (Stella and Klf4), and the increase in expression of markers of the primed state (Fgf5 and Dnmt3b) (Figure 3B). Immunostaining and western blot analyses further confirmed that naive cells that were grown in 2i/LIF express very low levels of DNMT3B and trimethylated histone H3 lysine 9 (H3K9me3), while primed cells showed a marked increase in these proteins (Figures 3C–3E). These data suggest that, in the presence of 2i/LIF, cells display marks of open chromatin, while in KSR/bFGF cells underwent a switch to primed state that is accompanied by epigenetic repressive marks. Importantly, the activity of all three RAS isoforms was significantly enhanced following the transition to the primed state, while the total level of the three RAS isoforms remained unchanged (Figure 3F). To further confirm that RAS activation is linked with activation of canonical RAS-downstream pathways, we examined the activity of the putative RAS effectors, AKT, ERK, JUN, and P38, using specific antibodies that recognize the phosphorylated (active) form of these proteins (i.e., pAKT, pERK, pJUN, and pP38), as well as the total levels of these proteins (AKT, ERK, JUN, and P38). Indeed, the activity of all of the above-mentioned RAS effectors, but not their actual level, was markedly induced in cells that were differentiated into the primed state (Figure 3G). To further explore if high RAS activity is general to cell priming, and not restricted to a specific culture condition, mESCs that were grown long term in FCS/LIF conditions were switched to primed (KSR/bFGF) state and harvested after 10 passages. Marker expression indicated that the cells were successfully differentiated from the naive to the primed state (Figures S4A–S4C). Importantly, here, too, the transition to the primed state was accompanied by an elevation in the relative activity of all RAS isoforms (Figure S4D). Altogether, these data confirmed that prominent RAS activation coincides with the transition from the naive (2i/LIF or FCS/LIF) to the primed state of mESCs. To further test if this correlation is relevant to human cells, we examined the levels of RAS-GTP in the transition between naive and primed PSCs in human cells. Naive and primed hESCs that were generated and maintained as described earlier (Gafni et al., 2013) and using qPCR showed correct marker expression (Figure 3H). Importantly, the levels of RAS-GTP significantly increase in primed cells (Figure 3I). Finally, to further examine whether this phenomenon is general and not restricted to a specific cell line, we examined the levels of RAS-GTP in different murine and human PSCs. Reassuringly, RAS-GTP levels were significantly higher in all primed cells, compared with naive cells while the total level of RAS remained unchanged, regardless of species or cell line specificity (Figure 3J).
Figure 3.
RAS Activity Is Induced in Primed ESCs
(A–G) mESCs that were grown in 2i/LIF were switched to KSR/bFGF and grown for ten passages. Bright-field images showing cell morphology of each state (A). Real-time qPCR (B), immunofluorescent staining (C and D), and western blot (E) analyses were performed to examine the relative expression of the indicated markers of naive (Stella and Klf4) or primed (Fgf5, Dnmt3b, and H3K9me3) state of pluripotency. DAPI is shown in (C) and (D). RAS-GTP pull-down assay was followed by western blot analysis using pan-RAS antibody (RAS-GTP) or using the indicated RAS isoform-specific antibodies (H-RAS-GTP, K-RAS-GTP, or N-RAS-GTP). The total expression of pan-RAS (RAS) or of each isoform (H-RAS, K-RAS, or N-RAS) was examined by western blot (F). The relative expression of active (phosphorylated) Ras-downstream effectors (pAKT, pERK, pJUN, and pP38) and the total expression was examined by western blot analysis using specific antibodies (G).
(H and I) Real-time qPCR (H) and RAS-GTP pull-down assay (I), followed by western blot analysis using pan-RAS antibody (RAS-GTP), were performed in naive and primed hESCs. AKT was used as a loading control.
(J) The levels of RAS-GTP, and total expression of pan-RAS and AKT (loading control), were examined in cell lysates from different mouse and human cell lines that were grown in 2i/LIF or in KSR/bFGF. mESC(1) = CGR8, mESC(2) = R1, hESC = H9, hiPSC(1) and hiPSC(2) were previously derived from skin fibroblast and hair follicles, respectively.
(E–G, I, and J) Values represent densitometry analysis of three independent experiments. Scale bars, 100 μm in (A), 50 μm in (C), and 5 μm in (D). Data shown are mean ± SD from three independent experiments. ∗p < 0.05 statistically significant by Student's t test.
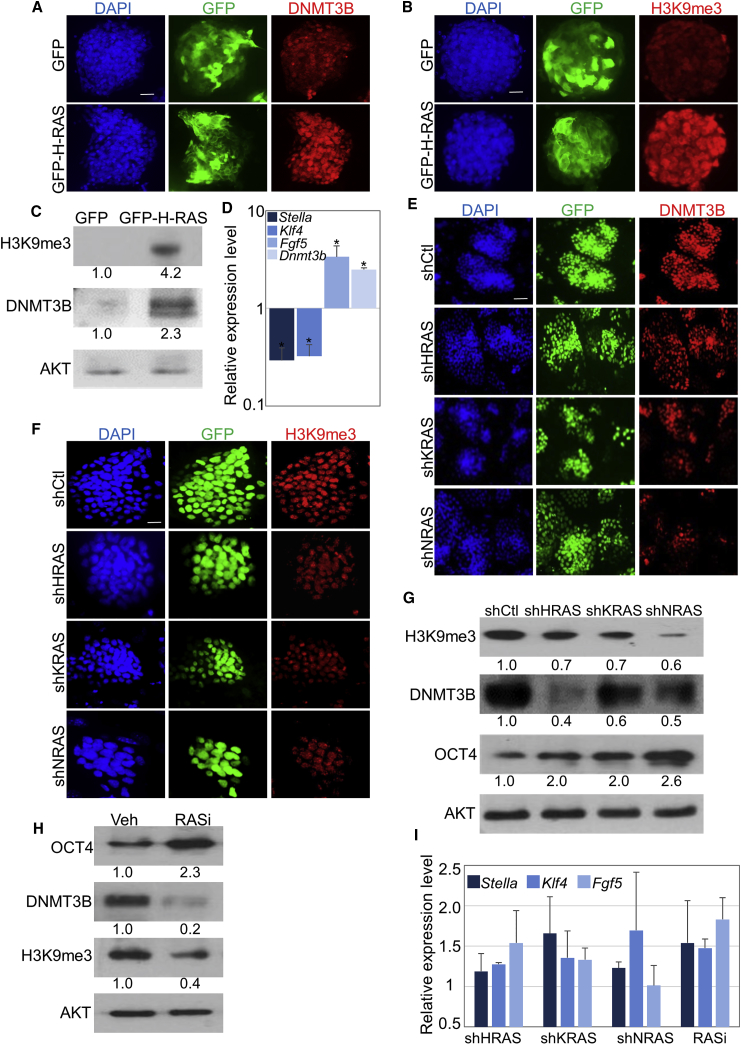
Next, we addressed the possibility that RAS can induce this process. Naive (2i/LIF) mESCs were transfected with GFP-H-RAS or GFP (as control), and harvested 48–72 hr later for analysis. Immunofluorescence and western blot analyses showed that RAS transfection induced a significant elevation in the levels of DNMT3B and H3K9me3 (Figures 4A–4C, S5A, and S5B). These data indicate that RAS expression alone is sufficient to induce a global repression pattern of the epigenetic landscape. Furthermore, real-time qPCR analysis revealed that markers of the naive state decreased, while markers of the primed state significantly increased in RAS-transfected cells (Figure 4D). These data indicate that forced expression of RAS alone is sufficient to destabilize naive pluripotency and induce cell differentiation. To further address this hypothesis in a more physiologically relevant manner, primed cells (KSR/bFGF) were infected with lentiviral vectors containing shRNAs against individual RAS isoforms, and DNMT3B and H3K9me3 were examined using immunofluorescence (Figures 4E, 4F, S5C, and S5D) and western blots (Figure 4G). In agreement, inhibition of each RAS isoform alone induced a partial but significant attenuation in histone methylation and DNMT3B expression (Figures 4E–4G, S5C, and S5D). Furthermore, we checked whether the depletion of all RAS isoforms by RASi showed similar results (Figure 4H). However, qPCR analysis showed that inhibition of individual RAS isoforms could not fully revert the expression pattern of the naive/primed markers (Figure 4I). This was also the case when primed cells were treated with RASi (Figure 4I). Thus, although RAS can induce differentiation (Figures 4A–4D), its inhibition in KSR/bFGF medium is not sufficient to fully revert cells to the naive state. Interestingly, the combination of RASi with MEK and GSKβ (2i) inhibitors resulted in a slight increase in the levels of Stella and Klf4 (but not Fgf5) (Figure S6). Altogether, these data strongly suggest that RAS is located in the hub of signaling pathways that control priming of naive cells and that it acts in concert with other pathways to control priming of PSCs.
Figure 4.
RAS Regulates the Transition from Naive to Primed ESCs
(A–D) mESCs grown in 2i/LIF medium were transfected with a plasmid encoding for GFP fused to H-RAS (GFP-H-RAS) or GFP alone. Cells were subjected to immunostaining (A and B), western blots (C), or real-time PCR (D) analyses of the indicated markers of naive (Stella and Klf4) or primed (Fgf5, DNMT3B, and H3K9me3) state.
(E–G) mESCs were grown in KSR/bFGF and infected with lentiviral vectors containing the indicated shRNAs against the different RAS isoforms together with nuclear GFP reporter. Immunofluorescence (E and F) and western blot analysis (G) of the indicated genes are shown.
(H) mESCs were grown in KSR/bFGF in the presence of the RASi (75 μM) for 48 hr following western blot analysis of the indicated genes.
(I) mESCs were grown in KSR/bFGF infected with the indicated shRNAs against specific Ras isoforms or in the presence of RASi (75 μM) for 72 hr. Real-time PCR analysis of the indicated genes is shown.
(C, G, and H) Values represent densitometry analysis of three independent experiments. Scale bar, 50 μm. Data shown are mean ± SD from three independent experiments. ∗p < 0.05 statistically significant by Student's t test.
Ras Regulates EMT and Changes in Metabolism during the Transition to the Primed State
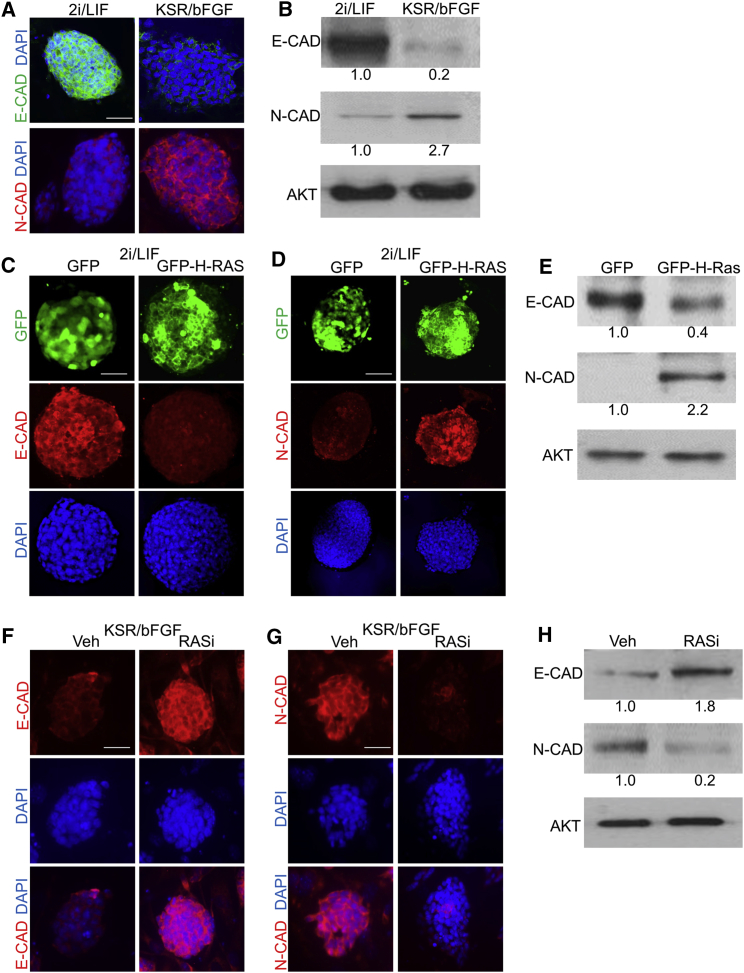
E-CADHERIN (E-CAD) is essential to maintain naive pluripotency (Faunes et al., 2013, Malaguti et al., 2013, Weinberger et al., 2016). In line with this, naive mESCs expressed high levels of E-CAD and low levels of N-CADHERIN (N-CAD), while primed cells displayed an opposite phenotype (Figures 5A and 5B). We hypothesized that RAS may regulate EMT during the transition to the primed state. Indeed, transfection of naive cells with GFP-H-RAS plasmid resulted in a marked reduction in E-CAD and an elevation in the levels of N-CAD (Figures 5C–5E and S7). In agreement, inhibition of RAS by RASi in primed cells significantly reduced N-CAD concomitant with an increase in E-CAD (Figures 5F–5H and S7). Altogether, these data indicate that RAS regulates the switch in CADHERIN expression in the naive-primed transition.
Figure 5.
RAS Controls the Switch in CADHERIN Expression in the Transition to the Primed State
(A and B) mESCs grown in 2i/LIF were switched to KSR/bFGF, grown for 10 passages, and subjected to western blot analysis (A) and immunofluorescent staining (B) of E-CADHERIN (E-CAD) or N-CADHERIN (N-CAD).
(C–E) mESCs were grown in 2i/LIF conditions and transfected with a plasmid encoding for GFP fused to H-Ras (GFP-H-RAS) or GFP alone. Immunostaining (C and D) or western blot analysis (E) of the indicated protein is shown.
(F–H) mESCs were grown in KSR/bFGF in the presence of RASi (75 μM) or Veh for 48 hr and then subjected to immunostaining or western blot analysis of the indicated CADHERIN.
(B, E, and H) Values represent densitometry analysis of three independent experiments. Scale bar, 50 μm.
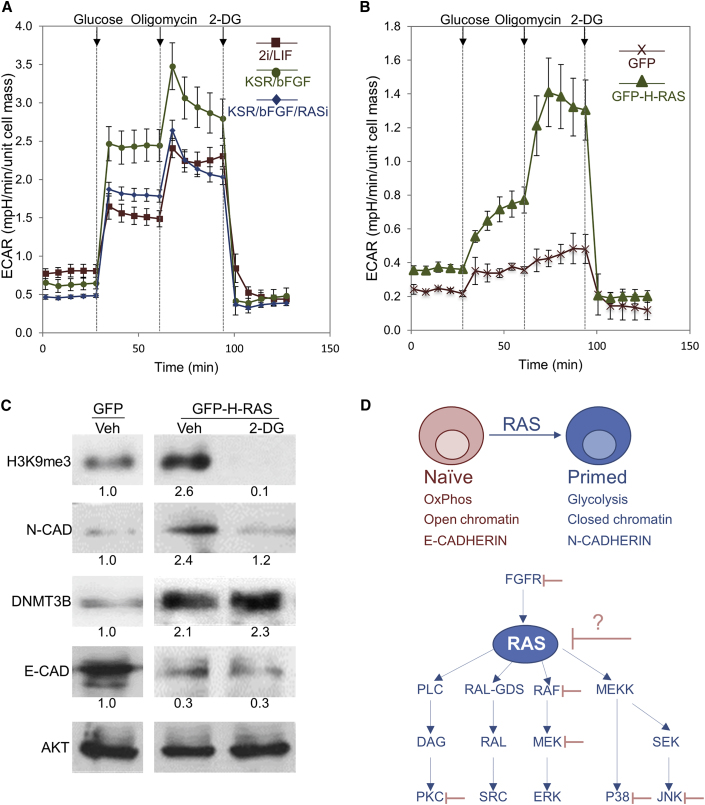
Finally, since the primed state is linked with increased glycolysis, we explored the role of RAS in controlling this metabolic switch during differentiation using the Seahorse extracellular flux analyzer. Extracellular acidification rate (ECAR) was recorded before and following the addition of glucose, oligomycin (inhibits ATP synthase), and 2-deoxyglucose (2-DG, inhibits glycolysis). As shown in Figure 6A, ECAR was significantly higher in primed cells compared with naive cells, indicative of increased glycolysis, which is associated with increased lactate production. Notably, 24 hr treatment of primed cells with RASi, resulted in a significant reduction in ECAR (Figure 6A), suggesting that RAS positively regulates glycolysis in primed cells. Moreover, RAS transfection in naive cells significantly enhanced ECAR, indicating that RAS plays a critical role in regulating metabolism during the naive-primed transition of mESCs (Figure 6B). Metabolic changes in naive-primed transition were linked with epigenetic changes and differentiation (Moussaieff et al., 2015). Notably, the effect of RAS on H3K9me3 and N-CAD was reversed when glycolysis was blocked by 2-DG (Figure 6C). By contrast, the RAS-mediated effect on DNMT3B and E-CAD was insensitive to 2-DG. Altogether, these data suggest that several RAS pathways are mediated by RAS-regulated metabolic cues while others are not.
Figure 6.
RAS Induces Glycolysis in mESCs
(A) mESCs were grown in the indicated media (2i/LIF or KSR/bFGF) or in KSR/bFGF in the presence of RASi (75 μM) or control (Veh). Extracellular acidification rate (ECAR) was recorded before and following the addition of glucose, oligomycin (inhibitor of ATP synthase which blocks OxPhos), or 2-deoxyglucose (2-DG, inhibitor of glycolysis).
(B) mESCs were grown in 2i/LIF conditions and transfected with a plasmid encoding for GFP fused to H-RAS (GFP-H-RAS) or GFP alone. ECAR was recorded before and following the addition of glucose, oligomycin, or 2-DG.
(C) mESCs were grown in 2i/LIF conditions and transfected with a plasmid encoding for GFP fused to H-RAS (GFP-H-RAS) or GFP as control. On the next day, 2-DG or Veh was added, and 48 hr later, cells were lysed and subjected to western blot analysis of the indicated proteins. AKT served as loading control. Values represent densitometry analysis of two independent experiments.
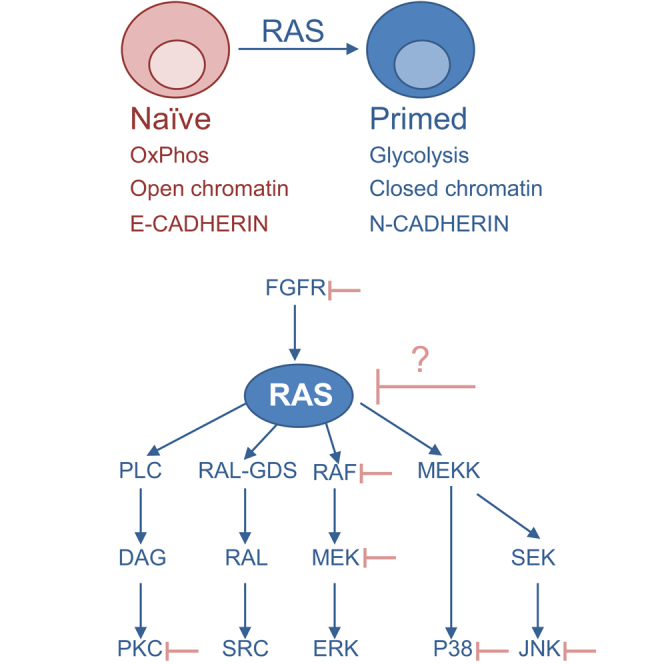
(D) Upper panel: schematic illustration of RAS function in the transition to the primed state. Lower panel: selected signaling pathways that are regulated by RAS, among them, several factors (FGFR, PKC, RAF, MEK, P38, and JNK) that are positioned upstream or downstream of Ras signaling pathway were targeted by pharmacological inhibitors to stabilize human cells in the naive state. We propose that, since RAS controls a large set of crucial signaling protein, its targeting may be useful approach for reprogramming human cells into naive state. Data shown are mean ± SD from three independent experiments.
Discussion
RAS proteins are ubiquitously expressed in many tissues where they control various processes including cell proliferation, migration, differentiation, and apoptosis (Mitin et al., 2005, Prior and Hancock, 2012). These diverse activities are transduced through the interaction with more than 20 factors that are known as RAS effector proteins (Schubbert et al., 2007, Vigil et al., 2010). RAS isoforms share many overlapping features and functions. Yet, an isoform-specific role (Yan et al., 1998) and an association with definite types of cancer have been reported (Prior et al., 2012). In the present study, RAS isoforms displayed a similar pattern of activation and overlapping roles in ESC differentiation. The three RAS isoforms play a redundant role in regulating an early exit from naive pluripotency to differentiation. The levels of active GTP-bound RAS proteins was strikingly low in naive cells. By contrast, activation of all three RAS isoforms was observed following differentiation that was induced through EBs formation, removal of LIF, or transitioning to primed state. This suggests that RAS plays a role in early development in vivo. The synchronized induction of RAS isoforms could result from receptor-mediated signaling, for example, via an FGF receptor.
Forced expression or inhibition of RAS revealed that RAS controls a range of events that are a hallmark of early differentiation of mESCs, including the increase in glycolysis, EMT, and epigenetic remodeling. The metabolic switch to anaerobic glycolysis is believed to be a central event in the transition from naive to primed state (Sperber et al., 2015, Wu and Belmonte, 2015). In line with the role of RAS in cancer cells (Cox et al., 2014), RAS regulates metabolism in ESCs. Inhibition of RAS decreased glycolysis in primed cells, while forced expression of RAS in naive cells increased glycolysis. Interestingly, RAS-mediated elevation in H3K9me3 was reversed when glycolysis was blocked by 2-DG, indicating that RAS-dependent metabolic pathways and global changes in the epigenetic landscape are linked, in line with previous reports (Sperber et al., 2015, Wu and Belmonte, 2015). While RAS-mediated upregulation of N-CAD was repressed by 2-DG, the regulation of DNMT3B and E-CAD by RAS was not affected by 2-DG. These data indicate that RAS exerts some aspects of its control on early differentiation through the control of metabolism. The regulation of metabolism by RAS, and its other functions, may be mediated by activation of assorted RAS effectors that have been implicated in the control of metabolism, epigenetics, and EMT in different cellular contexts, such as cancer (Cox et al., 2014, Pavlova and Thompson, 2016, Ying et al., 2012).
Upon the transition from naive to primed state, RAS activation was coupled with activation of a putative RAS downstream pathway, namely, PI3K/AKT, RAF/MEK/ERK, P38, and JUN (see Figure 6D). It might be interesting in future experiments to understand which of the RAS downstream pathways control the range events that were shown here to be mediated by RAS. Interestingly, the conversion of human primed cells into the naive state was facilitated by different protocols published by distinct groups (Chan et al., 2013, Gafni et al., 2013, Takashima et al., 2015, Theunissen et al., 2014, Ware et al., 2014). Intriguingly, the cocktail of inhibitors proposed by the different groups included factors that are known to be downstream of, or to interact with, RAS pathways. Among these factors were the FGF receptor inhibitor (Gafni et al., 2013) that is upstream of RAS, inhibitors of canonical RAS-downstream map kinases, RAF (Theunissen et al., 2014), MEK (Chan et al., 2013, Gafni et al., 2013, Takashima et al., 2015, Theunissen et al., 2014, Ware et al., 2014), P38 (Gafni et al., 2013), JNK (Gafni et al., 2013), and inhibitors of RAS-related signals, including PKC (Takashima et al., 2015) and SRC (Theunissen et al., 2014). Since RAS is positioned at the hub of cell signal transduction, and given that it tightly controls most of the above-mentioned cascades (Figure 6D), it is tempting to hypothesize that inhibition of RAS would bring fruitful results. Indeed, pharmacological inhibition of RAS by the RASi (farnesylthiosalicyclic acid, also known as FTS or Salirasib), or by specific shRNAs, resulted in attenuation and reversion of key hallmarks of differentiation. This was the case when differentiation was induced in the absence of LIF, or in the transition between naive to primed state. RAS therefore becomes a promising candidate to be further explored in cellular reprogramming and PSC regulation.
Further study of RAS isoforms and their downstream signals in hPSCs will provide a better understanding of pluripotency states and early embryonic development. Since RAS inhibition by RASi or shRNA did not fully convert primed cells into the naive state, indicating that RAS acts in concert with other signals in this process. It will be interesting to investigate the crosstalk between RAS signals and other putative pathways (e.g., LIF/STAT3, WNT/β-CATENIN, TGF/SMAD, and SRC) that may co-operate in the core regulatory circuitry of pluripotency, metabolism, and suppression of cell differentiation (Huang et al., 2015, Young, 2011). Finally, targeting RAS and/or RAS downstream pathways together with different pathways associated with differentiation may pave the way for optimal culture of human cells in ground state and efficient somatic cellular reprogramming.
Experimental Procedures
Tissue Culture and Differentiation
mESCs were grown in 2i/LIF medium (containing N2B27 [Ying et al., 2008], LIF [homemade], CHIR99021 [3 μM, PeproTech], and PD0325901 [1 μM, PeproTech]) were passed using trypsin. mESCs that were grown in FSC/LIF medium (containing 90% DMEM medium [Thermo Scientific], 10% FCS serum replacement [HyClone], 1 mM sodium pyruvate [Biological Industries], 1 mM non-essential amino acids [Biological Industries], 0.1 mM β-mercaptoethanol [Thermo Scientific], Pen-Strep solution [1:1,000, Biological Industries], and LIF [homemade]), were grown on a mitomycin C-treated mouse embryonic fibroblast (MEF) feeder layer. mESCs and iPSCs cell lines (previously characterized; Shalom-Feuerstein et al., 2013) that were grown in KSR/bFGF medium (containing 80% DMEM/F12 medium [1:1, Thermo Scientific:Biological Industries], 20% KSR [Gibco], 1 mM L-glutamine [Biological Industries], 1 mM non-essential amino acids [Biological Industries], 0.1 mM β-mercaptoethanol [Thermo Scientific], Pen-Strep solution [1:1,000, Biological Industries], and 10 ng/mL bFGF [PeproTech]) were passaged using collagenase type IV (1.5 mg/mL, Invitrogen). EBs differentiation was induced in differentiation medium (containing 80% DMEM medium [Thermo Scientific], 20% hiFBS [Biological Industries], 1 mM L-glutamine [Biological Industries], 1 mM sodium pyruvate [Biological Industries], 1 mM non-essential amino acids [Biological Industries], and 0.1 mM β-mercaptoethanol [Thermo Scientific]). Transfections with Xfect (Clontech) were performed according to the manufacturer's instructions. Lentivirus production was done as described previously (Dor-On et al., 2017). The following shRNAs were used: H-Ras gene NM_008284.2: target sequence, GTGAGATTCGGCAGCATAAAT; K-Ras NM_021284.6 gene target sequence, CTATACATTAGTCCGAGAAAT; N-Ras gene NM_010937.2 target sequence: CGAAAGCAAGTGGTGATTGAT. When necessary, GFP+ cells were sorted by a FACSAria (BD Biosciences).
Prior to differentiation through EBs or LIF removal, mESCs were grown on gelatin (0.1%)-coated dishes for one to two passages. To initiate EBs differentiation, cells were cultured in hanging drops in FCS/LIF medium lacking LIF for 3 days, then EBs were transferred to grow in suspension in differentiation medium for another 3–6 days. To initiate early exiting from self-renewal, cells were washed with PBS and transferred to FCS/LIF medium lacking LIF for 4 or 48 hr in the presence of the RASi farnesylthiosalicyclic acid (75 μM) or Veh (0.1% DMSO). For differentiation from naive to primed state of pluripotency, CGR8 cells that were routinely grown in 2i/LIF were transferred to MEFs co-cultured in KSR/bFGF medium for ten passages. Human naive cells were generated and maintained in conditioned RsET (STEMCELL Technologies) commercial media and grown as described previously (Gafni et al., 2013, Shahbazi et al., 2017).
Western Blot, RAS-GTP Pull-Down Assay, and Antibodies
Cells were lysed with whole-cell lysis buffer, and proteins were subjected to PAGE in the presence of SDS. Proteins were separated on 12% polyacrylamide gel and transferred to nitrocellulose membranes (Bio-Rad). The membranes were probed with one of the following antibodies: mouse α-Pan-RAS (1:1,500, Calbiochem, OP40), mouse α-K-RAS (1:100, Calbiochem, OP24), mouse α-H-RAS (1:80, Calbiochem, OP23), mouse α-N-RAS (1:100, Calbiochem, OP25), rabbit α-ERK (1:3,500, Santa Cruz Biotechnology, sc154), mouse α-pERK (1:1,000, Abcam, ab201015), rabbit α-AKT (1:3,500, Cell Signaling Technology, no. 9272), rabbit α-pAKT (1:200, Cell Signaling Technology, no. 9271), rabbit α-pP38 (1:1,000, Abcam, ab195049), rabbit α-P38 (1:1,000, Cell Signaling Technology, sc7972), rabbit α-pJUN (1:1,000, Cell Signaling Technology, no. 9164), rabbit α-c-Jun (1:1,000, Santa Cruz Biotechnology, sc45), rabbit α-GFP (1:5,000, Abcam, ab13970), rabbit α-NANOG (1:1,000, PeproTech, 500-P236), rabbit α-H3K9me3 (1:1,000, Abcam, ab8898), rabbit α-DNMT3B (1:1,000, Abcam, ab2851), rabbit E-CAD (1:1,000, Santa Cruz Biotechnology, sc7870), rabbit α-N-CAD (1:1,000, Abcam, ab18203), and rabbit α-OCT3/4 (1:2,000, Santa Cruz Biotechnology, sc9081) at 4°C, overnight. Membranes were washed and incubated with peroxidase-conjugated goat α-mouse immunoglogulin G (IgG) (1:3,000, Jackson Laboratory, 111-035-144) or peroxidase-conjugated goat α-rabbit IgG (1:3,000, Jackson Laboratory, 115-035-062). Protein bands were visualized with an ECL kit (Biological Industries) and quantified by ImageJ software.
RAS-GTP pull-down was performed as described previously (Frangioni and Neel, 1993, Fridman et al., 2000). In brief, glutathione-agarose beads (Sigma, G4510) were coated with recombinant protein chimera of glutathione S-transferase (GST) fused to the RAS-binding domain of RAF (RBD). Lysates were incubated with GST-RBD beads and precipitates were probed by western blot.
Immunofluorescent Staining and Real-Time qPCR Analysis
Cells that were grown on cover slips were fixed (paraformaldehyde [Sigma], 4%, for 15 min), then permeabilized (Triton X-100 [BioLab], 0.1%, for 10 min), blocked (BSA [Invitrogen], 2.5%, for 30 min), incubated with primary Abs (listed above, overnight at 4°C), washed and incubated with secondary antibody (Alexa Fluor 488 or Alexa Fluor 594 donkey α-rabbit IgG [Invitrogen, A21206 and A21209], 1:500, for 1 hr at room temperature). Nuclei were stained with DAPI (Sigma) and mounted (Thermo Scientific). Images were taken by a Nikon’s Eclipse NI-E upright microscope or a ZEISS LSM8 80 confocal system. Quantification of immunostaining was performed using NIS-Elements analysis D software. Five to ten different field from each experiment were imaged and quantified by the software.
RNA was isolated using TRI Reagent (Sigma) according to manufacturer's instructions. cDNA was prepared by RT-PCR using the high-capacity cDNA synthesis kit (Applied Biosystems) according to manufacturer's instructions. Real-time qPCR was performed with Fast SYBR green master mix (Thermo Scientific). qPCR primers used in this study are listed in Table S1. Samples were cycled using the StepOnePlus (Applied Biosystems) qPCR system. Relative gene expression was normalized to GAPDH and calculated according to the ΔΔCT method for qPCR.
Metabolic Assay
ECAR measurements were done as described previously (Zhou et al., 2012) with an optical fluorescent oxygen/hydrogen sensor XFe96 Seahorse analyzer. mESCs that were grown in 2i/LIF (100,000 cells) or in KSR/bFGF (40,000 cells) were seeded on lysed MEFs (0.5% Triton and 0.034% NH4OH). The glycolysis stress kit was used to measure ECAR response using following addition of glucose (10 mM), oligomycin (2 μM), and 2-DG (50 mM). The data were normalized by modified Lowery method to quantify protein content after metabolite extraction.
Statistical Analysis
Data are presented as mean ± SEM. Comparison between means was evaluated using paired t test. ∗ indicates a p value of <0.05 and this value was considered to be statistically significant.
Author Contributions
A.A. was involved in conceptual and experimental design, performed and interpreted the experiments, prepared the figures, and participated in the manuscript writing; M.L., S.B., and I.A. designed and performed experiments, analyzed data, and prepared the figures; E.G. and J.H.H. were involved in conceptual and experimental design, data analysis, and manuscript writing; R.H. and Y.K. provided reagents and materials; R.S.-F. was involved in conceptual and experimental design, data interpretation, and manuscript writing. All authors approved the manuscript.
Acknowledgment
We thank Prof. Eran Meshorer for providing Rex1-YFP cells and for critical reading of the manuscript and Dr. Victoria Makovski and the interdisciplinary unit of our faculty for the technical support. The research leading to these results (R.S.-F.) has received funding from the European Union's Seventh Framework Program (FP7/2007-2013) under grant agreement no. 618432-MC-Epi-Patho-Stem and from the Ministry of Science, Technology and Space, Israel, the Ministere de L'Education National de L'Enseignement Supérieur de la Recherche (3-11985), the Israel Science Foundation, and the Bruce & Ruth Rappaport institute.
Published: February 15, 2018
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.004.
Supplemental Information
References
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. Developmental cell the ability of inner cell mass cells to self-renew as embryonic stem cells is acquired upon epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T., Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustinza-Linares E., Kurzrock R., Tsimberidou A.M. Salirasib in the treatment of pancreatic cancer. Futur. Oncol. 2010;6:885–891. doi: 10.2217/fon.10.71. [DOI] [PubMed] [Google Scholar]
- Chan Y.S., Göke J., Ng J.H., Lu X., Gonzales K.A.U., Tan C.P., Tng W.Q., Hong Z.Z., Lim Y.S., Ng H.H. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor-On E., Raviv S., Cohen Y., Adir O., Padmanabhan K., Luxenburg C. T-plastin is essential for basement membrane assembly and epidermal morphogenesis. Sci. Signal. 2017;10:1–9. doi: 10.1126/scisignal.aal3154. [DOI] [PubMed] [Google Scholar]
- Faunes F., Hayward P., Descalzo S.M., Chatterjee S.S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A.K., Dasgupta R. A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 2013;140:1171–1183. doi: 10.1242/dev.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni J.V., Neel B.G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Fridman M., Maruta H., Gonez J., Walker F., Treutlein H., Zeng J., Burgess A. Point mutants of c-Raf-1 RBD with elevated binding to v-Ha-Ras. J. Biol. Chem. 2000;275:30363–30371. doi: 10.1074/jbc.M003193200. [DOI] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A.W., Cassady J.P., Staerk J., Carey B.W., Lengner C.J., Foreman R., Love J. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Ye S., Zhou X., Liu D., Ying Q.L. Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell. Mol. Life Sci. 2015;72:1741–1757. doi: 10.1007/s00018-015-1833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Smith A. Mapping the route from naive pluripotency to lineage specification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0540. 20130540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguti M., Nistor P.A., Blin G., Pegg A., Zhou X., Lowell S. Bone morphogenic protein signalling suppresses differentiation of pluripotent cells by maintaining expression of E-Cadherin. Elife. 2013;2:e01197. doi: 10.7554/eLife.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitin N., Rossman K.L., Der C.J. Signaling interplay in Ras superfamily function. Curr. Biol. 2005;15:563–574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I.A., Hancock J.F. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Schubbert S., Shannon K., Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Shahbazi M.N., Scialdone A., Skorupska N., Weberling A., Recher G., Zhu M., Jedrusik A., Devito L.G., Noli L., Macaulay I.C. Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature. 2017;552:239–243. doi: 10.1038/nature24675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Feuerstein R., Lindenboim L., Stein R., Cox A.D., Kloog Y. Restoration of sensitivity to anoikis in Ras-transformed rat intestinal epithelial cells by a Ras inhibitor. Cell Death Differ. 2004;11:244–247. doi: 10.1038/sj.cdd.4401334. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R., Cooks T., Raz A., Kloog Y. Galectin-3 regulates a molecular switch from N-Ras to K-Ras usage in human breast carcinoma cells. Cancer Res. 2005;65:7292–7300. doi: 10.1158/0008-5472.CAN-05-0775. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R., Plowman S.J., Rotblat B., Ariotti N., Tian T., Hancock J.F., Kloog Y. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 2008;68:6608–6616. doi: 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Feuerstein R., Serror L., Aberdam E., Muller F.J., van Bokhoven H., Wiman K.G., Zhou H., Aberdam D., Petit I. Impaired epithelial differentiation of induced pluripotent stem cells from ectodermal dysplasia-related patients is rescued by the small compound APR-246/PRIMA-1MET. Proc. Natl. Acad. Sci. USA. 2013;110:2152–2156. doi: 10.1073/pnas.1201753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K.A., Devi A., Detraux D., Gu H. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Erratum: resetting transcription factor control circuitry toward ground-state pluripotency in human (Cell (2014) 158 (1254-1269)) Cell. 2015;162:452–453. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D., Cherfils J., Rossman K.L., Der C.J. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C.B., Nelson A.M., Mecham B., Hesson J., Zhou W., Jonlin E.C., Jimenez-Caliani A.J., Deng X., Cavanaugh C., Cook S. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Weisz B., Giehl K., Gana-Weisz M., Egozi Y., Ben-Baruch G., Marciano D., Gierschik P., Kloog Y. A new functional Ras antagonist inhibits human pancreatic tumor growth in nude mice. Oncogene. 1999;18:2579–2588. doi: 10.1038/sj.onc.1202602. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Smith A.G. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Wu J., Belmonte J.C. Metabolic exit from naive pluripotency. Nat. Cell Biol. 2015;17:1519–1521. doi: 10.1038/ncb3269. [DOI] [PubMed] [Google Scholar]
- Yan J., Roy S., Apolloni A., Lane A., Hancock J.F. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu B., Xu J., Wang J., Wu J., Shi C., Xu Y., Dong J., Wang C., Lai W. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell. 2017;169:243–257.e25. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.