Figure 5.
Nutlin-3 Promotes Reserve Cell Generation in Serum-Deprived Myoblasts
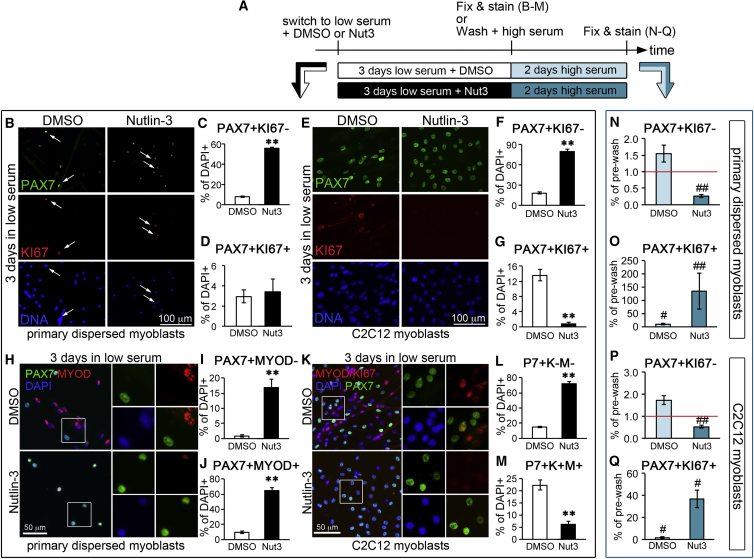
(A) Schematic representation of the reserve cell assay experimental design. Primary and C2C12 myoblast cultures were switched to low serum medium to induce cell-cycle exit in the presence of either 20 μM Nutlin-3 (Nut3) or vehicle (DMSO). To distinguish between quiescence and senescence, myoblast cultures that had been maintained in low serum supplemented with either DMSO or Nutlin-3 for 3 days were re-exposed to high serum for 2 days. The experiment was repeated 3 times independently and each time 10–15 technical replicates were scored.
(B–D) Primary myoblast cultures were treated as in (A) and, after 3 days in low serum, fixed, immunostained to detect PAX7 (green), KI67 (red), and DNA (DAPI, blue), and scored for the percentages of PAX7+/KI67– (C) and PAX7+/KI67+ (D) cells. In (B), one representative image for each treatment is shown. In (C) and (D) quantitative analyses of the indicated cell subpopulations across all three independent experiments were plotted as average ± SEM. The arrows in (B) indicate PAX7+/KI67+ cells.
(E–G) C2C12 myoblasts cultures were treated as in (A) and, after 3 days in low serum, fixed, and immunostained as in (B). In (E) one representative image for each treatment is shown. In (F) and (G) quantitative analyses of the indicated cell subpopulations across all three independent experiments were plotted as average ± SEM.
(H–M) Primary (H and J) and C2C12 (K–M) myoblasts that had been maintained in low serum supplemented with either DMSO or Nutlin-3 for 3 days were fixed and immunostained to detect PAX7 (green), KI67, and/or MYOD (red) and DNA (DAPI, blue). In (H) and (K) one representative image for each treatment is shown. Insets are enlarged on the side of the main image to show mutual exclusion or co-localization of PAX7 and MYOD (H) or PAX7 and MYOD+ KI67 (K). In (I), (J), (L), and (M) quantitative analyses of the indicated cell subpopulations across all three independent experiments were plotted as average ± SEM.
(N–Q) In order to quantify true reserve cells, after 3 days in low serum, primary (N and O) and C2C12 (P and Q) myoblast cultures were re-exposed to high serum as depicted in (A) for 2 days, then fixed and immunostained to detect PAX7, KI67, and DNA. The percentages of PAX7+/KI67– (N and P) and PAX7+/KI67+ (O and Q) were calculated and plotted as fold change of the same population in cultures that had been maintained in low serum and then re-exposed to high serum (we will call it “post-wash” for simplicity) versus the same population in cultures maintained for only 3 days in low serum (indicated as “pre-wash”). The average of the post-wash/pre-wash fold change for each subpopulation across 3 independent biological replicates (each one scored for 10–15 technical replicates) was calculated and plotted ± SEM.
In (C) and (D), (F) and (G), (I) and (J), and (L) and (M): ∗∗p < 0.01, where p is the p value of the average percentage of each cell subpopulation in Nut-3-treated cultures versus DMSO-treated cultures. In (N)–(Q), #p < 0.05 and ##p < 0.01, where p is the p value of the fold change calculated as “subpopulation percentage in post-wash/subpopulation percentage in pre-wash” within each treatment (DMSO or Nut-3).