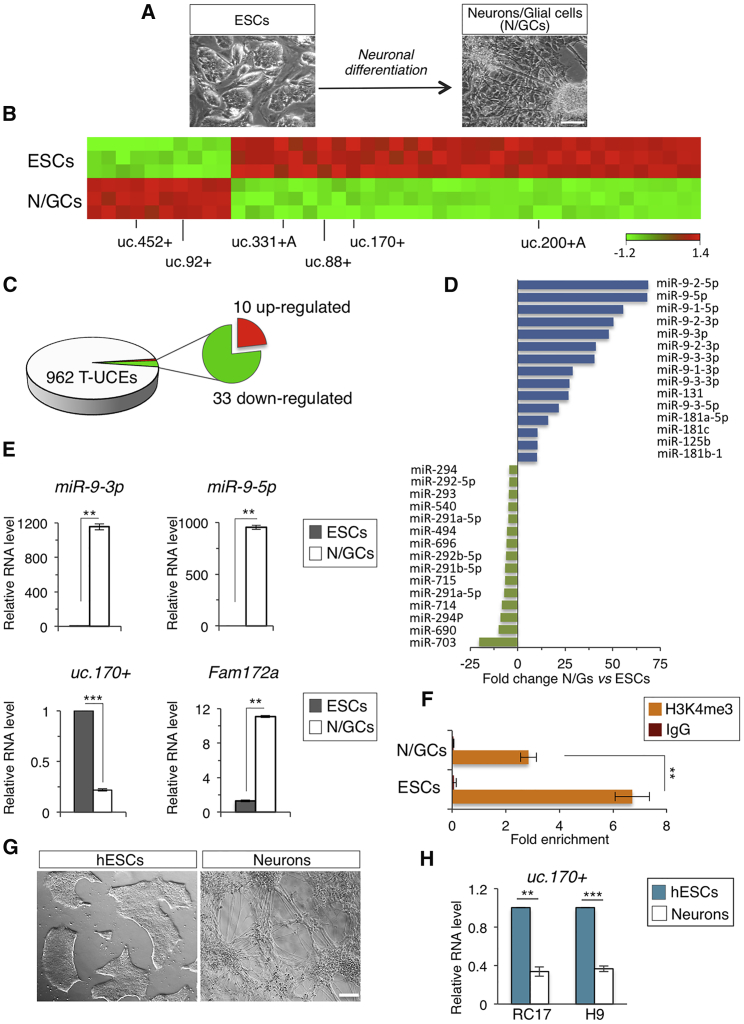
Figure 1.
Genome-wide Expression Profiling of T-UCEs and miRNAs in ESC Differentiation
(A) Schematic representation and representative photomicrographs of the ESC neural differentiation. Scale bar, 100 μm.
(B) Heatmap diagram of differentially expressed sense (+) and antisense (+A) T-UCEs, in ESC-derived neurons/glial cells (N/GCs) versus undifferentiated cells (ESCs) (p < 0.001; two-sample t test). Randomly selected T-UCEs are indicated among all the deregulated.
(C) Microarray-based pie chart representing global distribution of differentially expressed T-UCEs in N/GCs versus ESCs.
(D) Bar plot showing the differentially expressed miRNAs in N/GCs versus ESCs with the largest change in expression (p < 0.001; two-sample t test).
(E) qRT-PCR analysis of miR-9-5p/3p, uc.170+, and Fam172a in ESCs and N/GCs. Relative RNA level was normalized to either Gapdh or U6 for coding/non-coding genes, respectively. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(F) ChIP-qPCR of H3K4me3 at uc.170+ (ultraconserved region) locus in ESCs and N/GCs. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.01.
(G) Representative photomicrographs of the human ESC neural differentiation. Scale bar, 200 μm.
(H) qRT-PCR analysis of uc.170+ in human ESCs and human neurons (RC17 and H9 are two different human ESC lines). Relative RNA level was normalized to U6. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001. See also Figure S1.