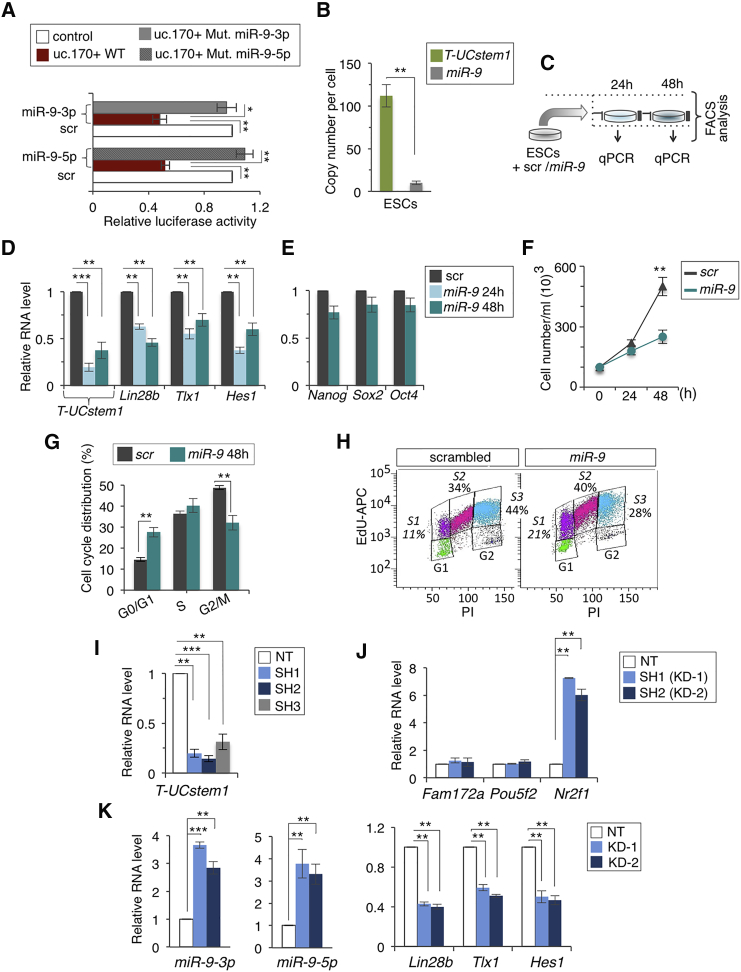
Figure 2.
Functional Interaction of T-UCstem1 and miR-9 in ESCs
(A) Luciferase reporter assay with Renilla luciferase under control of uc.170+ sequence wild-type (WT) or mutant (Mut) co-transfected with miR-9-5p/3p or scrambled (scr) miRNA (100 nmol) in 293FT cells. The luciferase activity of Firefly was used as internal control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.01, ∗∗p < 0.005.
(B) T-UCstem1 and miR-9 copy number per cell quantified with qRT-PCR in ESCs. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005.
(C) Schematic representation of the experimental procedure.
(D and E) qRT-PCR analysis of (D) T-UCstem1, miR-9 targets (Lin28b, Tlx1, and Hes1), and (E) pluripotency-associated genes (Nanog, Sox2, and Oct4) in ESCs transfected with miR-9 or scr (100 nmol) for 24 and/or 48 hr. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(F) Automated cell counting of miR-9 and Control (scr) transfected ESCs at 24 and/or 48 hr. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005.
(G and H) FACS-based cell-cycle distribution analysis of ESCs transfected with miR-9 or miRNA scr for 48 hr (G) after PI or (H) after double 5-ethynyl-2′-deoxyuridine (EdU)/propidium iodide (PI) staining (representative FACS plots of biological duplicates are shown). ∗∗p < 0.005.
(I and J) qRT-PCR analysis of (I) T-UCstem1, (J) host and neighbor genes (Fam172a, Pou5f2, and Nr2f1) in NT and independent T-UCstem1 KD ESC clones. Relative RNA level was normalized to either Gapdh or U6 for coding/non-coding genes, respectively. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001.
(K) qRT-PCR analysis of miR-9-3p/5p and its target genes, Lin28b, Tlx1, and Hes1. Relative RNA level was normalized to either Gapdh or U6 for coding/non-coding genes, respectively. Data are mean ± SEM (n = 3 independent experiments); ∗∗p < 0.005, ∗∗∗p < 0.001. See also Figures S1 and S2.