Figure 3.
T-UCstem1 Depletion Affects ESC Cell Proliferation and Induces Apoptosis
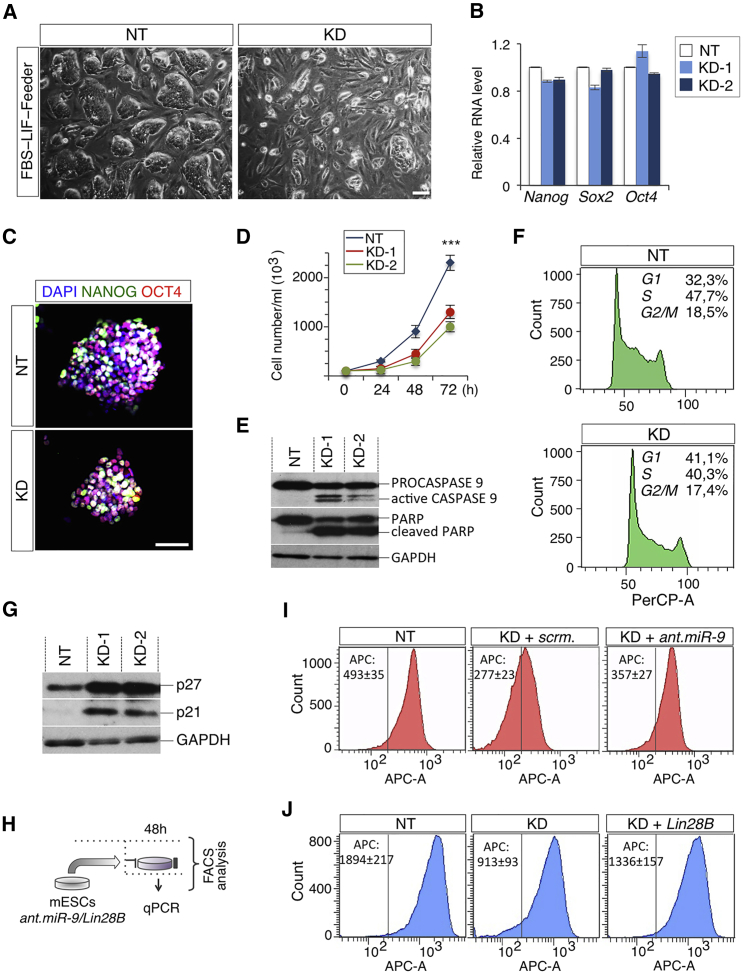
(A) Representative photomicrographs of FBS/LIF/Feeders non-targeted (NT) and T-UCstem1 KD (KD) ESCs. Scale bar, 100 μm.
(B) qRT-PCR of pluripotency genes (Nanog, Sox2, and Oct4) in Control and two independent KD ESC clones. Relative RNA level was normalized to Gapdh. Data are mean ± SEM (n = 3 independent experiments).
(C) Representative pictures of OCT4/NANOG double immunostaining of NT and KD ESCs. Nuclei were stained with DAPI. Scale bars, 75 μm.
(D) Time course analysis of automated cell counting of FBS/LIF/Feeders Control (NT) and KD ESCs. Data are mean ± SEM (n = 3 independent experiments); ∗∗∗p < 0.001.
(E) Western blot analysis of PROCASPASE/active CASPASE9 and PARP full-length form and cleaved fragment in Control (NT) and two independent T-UCstem1 KD ESC clones. GAPDH was used as loading controls.
(F) Cell-cycle distribution by PI incorporation in Control and KD ESCs. Representative FACS plots of biological triplicates are shown.
(G) Western blot analysis of cell-cycle inhibitors (p27and p21) in Control (NT) and two independent T-UCstem1 KD ESC clones. GAPDH was used as a loading control.
(H) Schematic representation of the experimental procedure.
(I and J) FACS-based analysis of cell proliferation quantification by EdU incorporation in NT and in KD ESCs upon antagomiR-9 5p/3p/scr (100 nmol) (I) or Lin28B-Flag/empty vector at 48 hr after transfection (J). Representative FACS plots of biological triplicates are shown. Data are mean ± SEM. See also Figure S3.