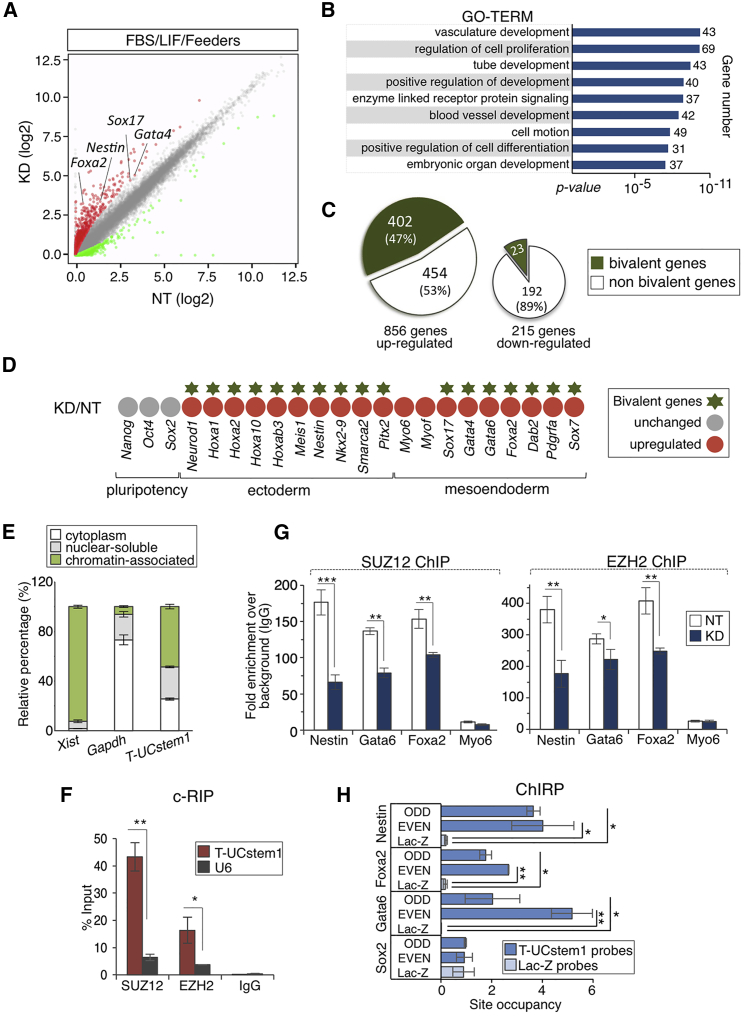
Figure 6.
T-UCstem1 Depletion Remodels the Epigenomic Signature of ESCs
(A) Scatterplot of RNA-seq data shows differentially expressed genes in T-UCstem1 KD (KD) versus Control (NT) ESCs. The gray circles delineate the boundaries of less than 2-fold difference in gene expression levels. Genes showing a significant (p < 0.05) higher or lower expression level in KD versus NT ESCs are indicated as red and green circles, respectively. Three independent KD ESC clones were analyzed and the average values were reported in the plot.
(B) Gene ontology (GO) (http://david.abcc.ncifcrf.gov; setting as background the genes expressed in ESCs) of protein coding genes deregulated in KD versus Control (NT) ESCs.
(C) Chart showing a significant overlap between bivalent domain-associated genes and upregulated genes in KD versus Control (Hypergeometric test; p <10−16).
(D) Heatmap of upregulated developmental genes in KD cells versus Control (NT).
(E) Total RNA from ESCs was separated into cytoplasmic, nuclear-soluble, and chromatin-bound fractions. The relative abundance of T-UCstem1 in the different fractions was measured by qRT-PCR. Gapdh and Xist were analyzed as control. Data are mean ± SEM (n = 3 independent experiments).
(F) Crosslinked RNA immunoprecipitation (c-RIP) of T-UCstem1 in ESCs, using antibodies against SUZ12, EZH2, or immunoglobulin (IgG) as control. U6 was analyzed as negative control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.05, ∗∗p < 0.005.
(G) ChIP-qPCR of SUZ12 and EZH2 binding at selected genes in NT and KD cells. Myo6 promoter has been reported as control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
(H) ChIRP-qPCR for T-UCstem1 in ESCs. Enrichments of Nestin, Gata6, Foxa2, and Sox2 promoter regions were quantified in the chromatin fraction precipitated using biotinylated DNA probes complementary to T-UCstem1 and LacZ as negative control. Sox2 promoter has been reported as negative control. Data are mean ± SEM (n = 3 independent experiments); ∗p < 0.05, ∗∗p < 0.005. See also Figure S6.