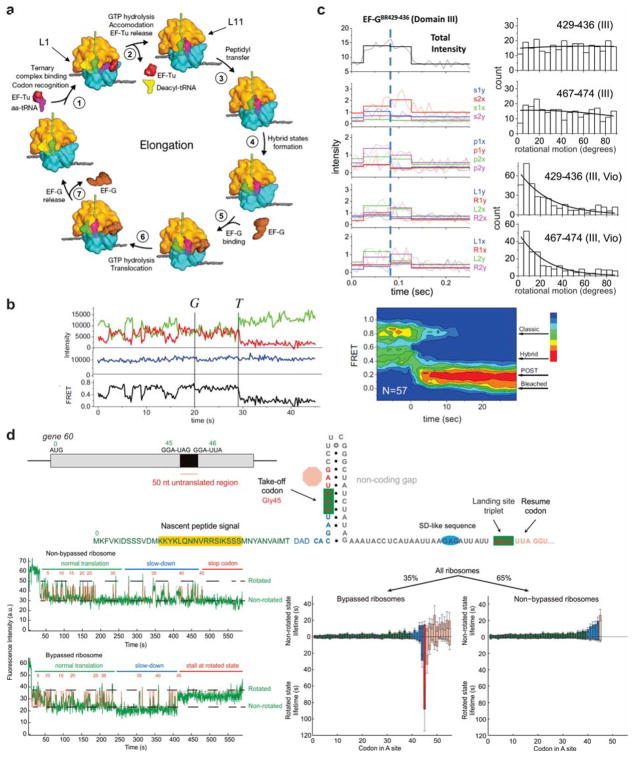
Figure 13. Protein and RNA dynamics during mRNA translation.
(a) The elongation cycle of the bacterial ribosome. (b) Study of ribosome translocation by monitoring FRET between multiple tRNAs and between tRNAs and the ribosome. In the absence of EF-G, the ribosome fluctuates between conformations in which the tRNAs are in classical (high-FRET) and hybrid (mid-FRET) states. Addition of EF-G (at the time marked “G” in the traces) suppresses these fluctuations, and leads to a rapid transition to a low-FRET post-translocation state. Panel (a) and (b) are reproduced with permission from Ref 278 Copyright 2011 Elsevier Inc. (c) Study of rotational motions in EF-G by polarization-resolved single-molecule microscopy. Left: rapid increases in overall fluorescence intensity indicate EF-G binding events, while changes in the relative intensities of 16 polarization-resolved signals indicate rotations of the labeled domain of EF-G. Right: Histograms binning the angles of rotations observed during EF-G binding events. Two different fluorophore positions (residues 429–436 and 467–474 of domain III) indicate that domain III exhibits large re-orientations. These re-orientations are suppressed in the presence of Viomycin. Reproduced with permission from Ref 279. (d) Translational bypassing on T4 gene 60 mRNA. Translating ribosomes fluctuate between rotated and non-rotated states. Upon reaching the “take-off” codon, ribosomes that bypass exhibit a long-lived rotated state. Reproduced with permission from Ref 280 Copyright 2015 Elsevier Inc.