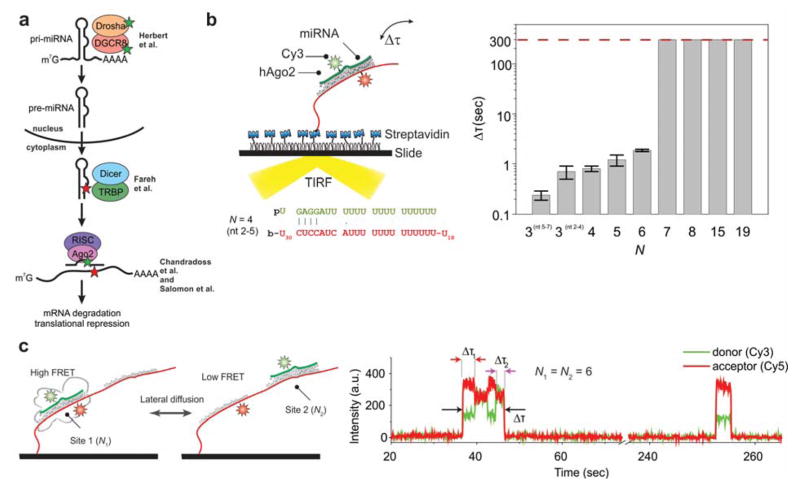
Figure 8. Recognition of RNA interference targets by Ago2.
(a) The miRNA biogenesis pathway. Colored stars indicate the labeling sites used for various studies discussed in the text. (b) Left: Binding of Ago2 loaded with fluorophore-labeled miRNA to a labeled, immobilized RNA target. Right: The bound time Δτ was determined as a function of the number of miRNA-target base pairs. It increases greatly when a 7-nucleotide seed sequence at the 5′ end of the miRNA is fully paired. (c) 1D diffusion of Ago2 was studied using an immobilized RNA target with two adjacent binding sites (left). A high-FRET state is observed when Ago2 is bound at “Site 1”, and a mid-FRET state is observed when it is bound at “Site 2” (right). Ago2 was found to fluctuate between the two binding sites without dissociating, implicating sliding as a mechanism for target location. Reproduced with permission from Ref 219 Copyright 2015 Elsevier Inc.