Fig. 4.
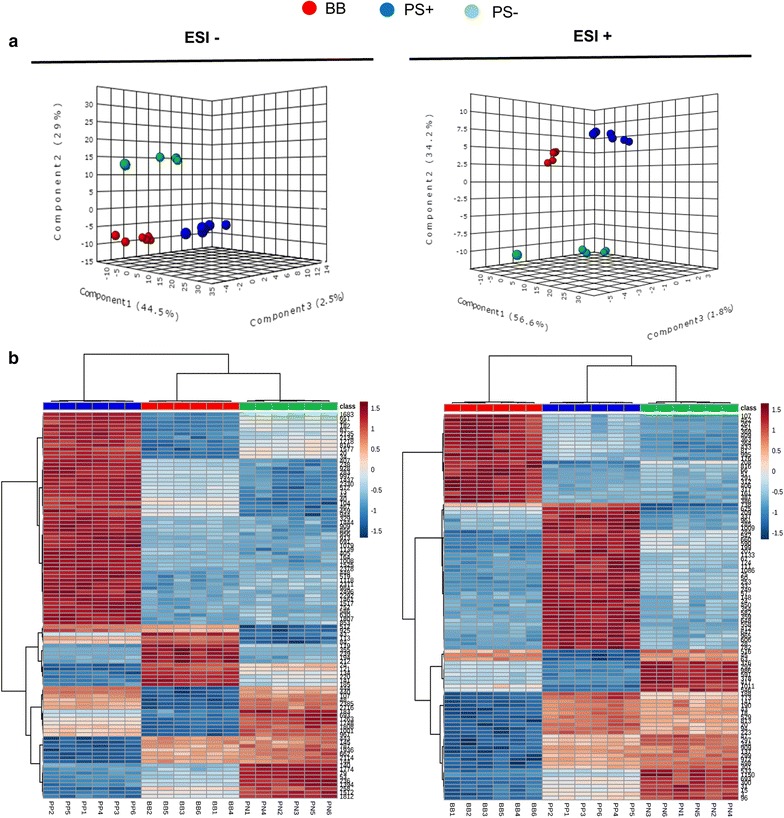
General outlook of the metabolic behaviour of the three genotypes of tomato plants using partial least squares, heatmap and clustering analysis. a 3D partial least squares plot explaining the major sources of variability for ESI (−) and ESI (+) signals obtained from a non-targeted analysis by UHPLC-QTOF MS in the three conditions BB, PS+ and PS−. Data points represent replicates of three independent experiments with two technical replicates per experiment and condition, which were injected randomly in the UHPLC-QTOF MS. Signals corresponding to different conditions were compared using the non-parametric Kruskal–Wallis test, and only data corresponding to p < 0.05 between groups were used for supervised analysis. b Heat map analysis and clustering of the different main signals corresponding to metabolite profiling, and generated with the Metaboanalyst 3.0 software, following a Kruskal–Wallis test (p < 0.05) in both ESI (−) and ESI (+) modes of ionization
