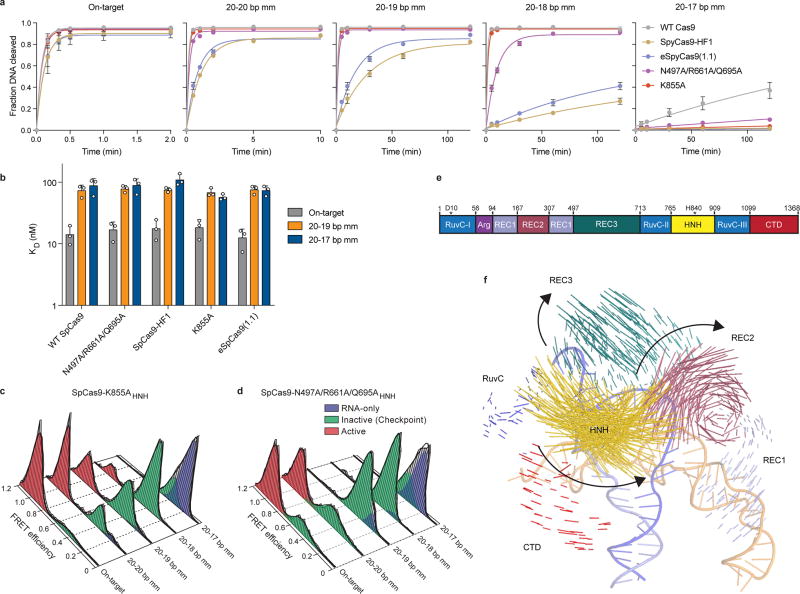
Extended Data Figure 2. HNH domain in eSpCas9 variants still populate the docked state in the presence of PAM-distal mismatches.
a, Quantification of DNA cleavage time courses comparing WT SpCas9, SpCas9-HF and eSpCas9(1.1) variants with perfect and PAM-distal mismatched targets. b, Dissociation constants comparing WT SpCas9, SpCas9-HF and eSpCas9(1.1) variants with perfect and PAM-distal mismatched targets, as measured by electrophoretic mobility shift assays. For panels a–b, mean and s.d. shown; n = 3 independent experiments (overlaid as white circles in panel b). c–d, smFRET histograms for c, SpCas9-K855A and d, SpCas9-N497A/R661A/Q695A. For panels c and d, black curves represent a fit to multiple Gaussian peaks. e, Schematic of SpCas9 domain structure with color coding for separate domains. f, Vector map of global SpCas9 conformational changes from the sgRNA- (PDB ID: 4ZT0) to dsDNA-bound structures (PDB ID: 5F9R), domains colored as in panel e.