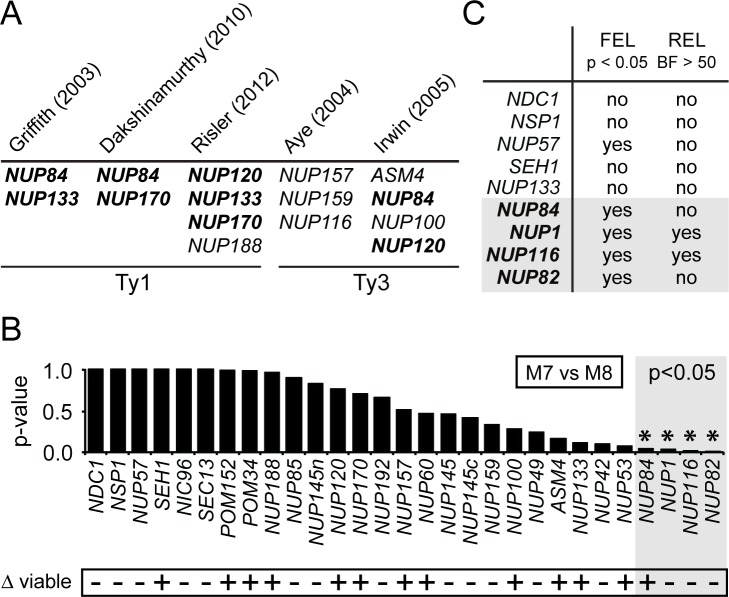
Fig 2. Nucleoporins are evolving rapidly in Saccharomyces yeasts.
(A) Results of published high-throughput genetic screens for host factors affecting Ty mobility [51,52,55–57]. Only nucleoporin genes found in these screens are summarized, where disruption of the indicated gene altered Ty3 or Ty1 mobility. Bold text indicates genes found in more than one screen. (B) Results from PAML analysis surveying nucleoporin genes for codons with elevated evolutionary rate (dN/dS ≥1). Here, alignments were fit to a codon model of conservative evolution (M7) and a codon model allowing for codons with an elevated evolutionary rate (M8). M7 was rejected in favor of M8 for four nucleoporins (p<0.05): NUP84, NUP1, NUP116 and NUP82. Along the bottom is summarized whether yeast with a deletion of each of these genes is viable, taken from the Saccharomyces genome database. (C) Extended evolutionary analysis of selected nucleoporins using two additional tests for positive selection (FEL and REL) [68]. “Yes” indicates that codons with dN/dS>1 were detected in this gene by the indicated test, with a p-value (p) < 0.05, or Bayes factor (BF) > 50.