Figure 6.
Gene Expression Profiling of FLK-1+PDGFRα− Cells
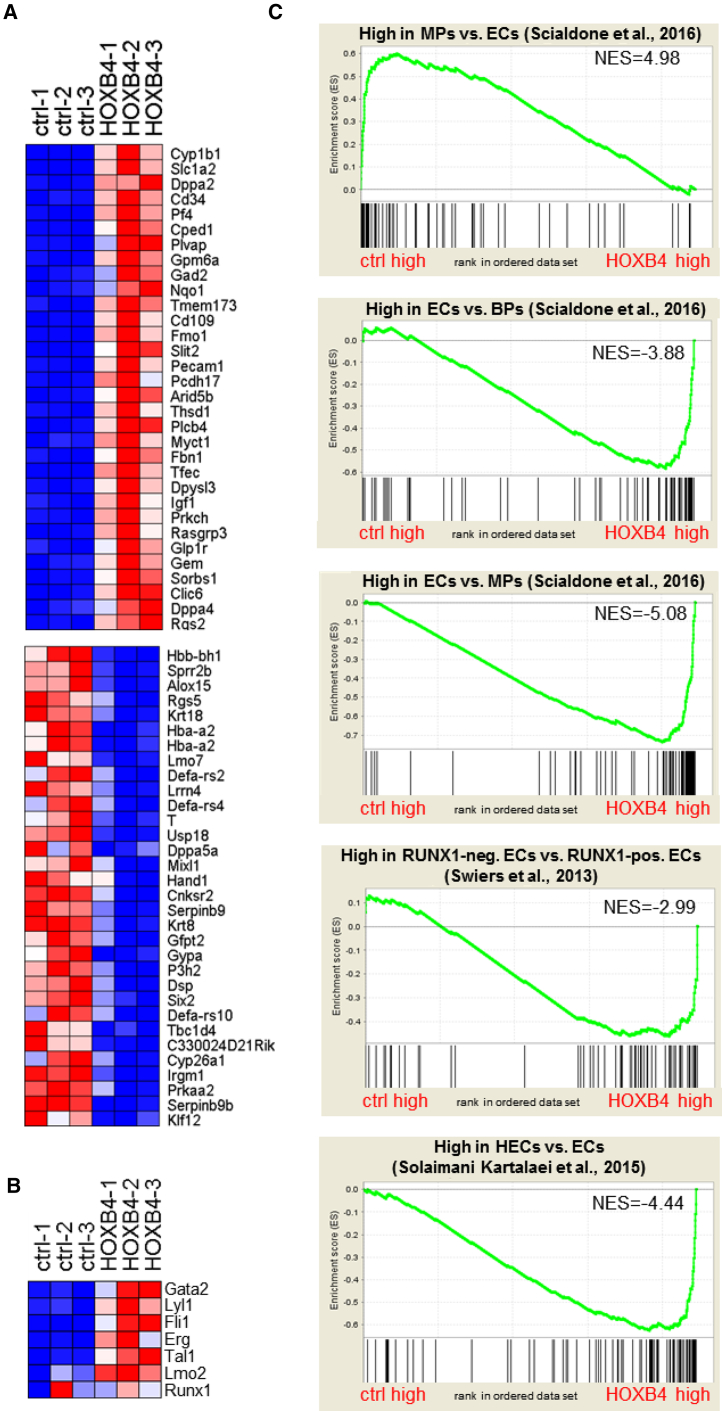
(A) Heatmap showing the top 32 genes (fold-change ranked) either up- or downregulated in FLK-1+PDGFRα− cell populations from iRunx day 4 EBs without (ctrl) or with ectopically expressed HOXB4. Genes were considered differentially expressed if they met two criteria: q ≤ 0.2 and fold-change ≥ 2.0 (499 genes) (for full list see Table S1).
(B) Heatmap for the expression of heptad transcription factors.
(C) GSEA using gene sets specific for mesodermal progenitors ([MPs] top panel) compared with endothelial cells (ECs), ECs compared with blood progenitors ([BPs] second panel) and endothelial cells compared with mesodermal progenitors (third panel) (Scialdone et al., 2016); E8.5 RUNX− endothelial cells (RUNX1-neg. ECs) compared with RUNX1+ endothelial cells (RUNX1-pos. ECs, fourth panel) (Swiers et al., 2013) and E10 hemogenic endothelial cells (HECs) compared with endothelial cells (ECs, fifth panel) (Solaimani Kartalaei et al., 2015). Genes were drawn according to their rank from left (high expression in control) to right (high expression in HOXB4-expressing cells) and gene sets plotted on top with each black bar representing a gene. The enrichment score is plotted in on the vertical axis.