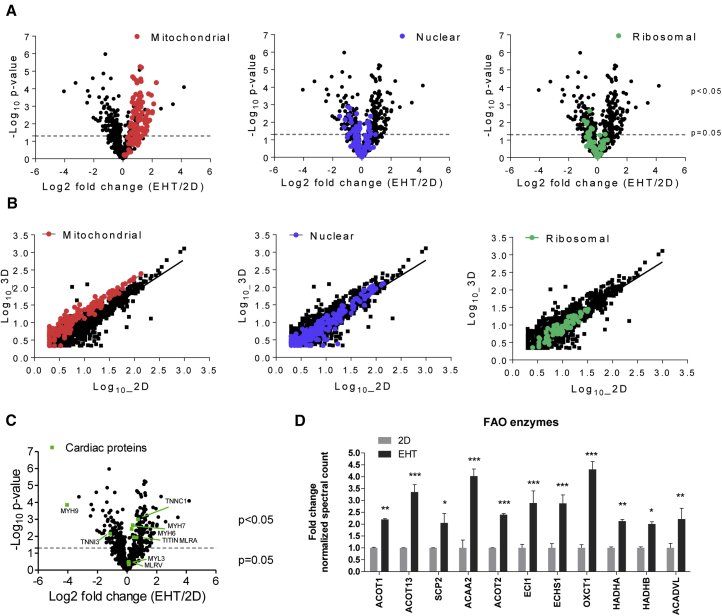
Figure 2.
Quantitative Analysis of Proteomic Profiles of Human iPSC-Derived 2D or EHT Samples
(A) Volcano plots to compare the mean log2 fold changes (EHT/2D) of normalized spectral counts and the log10 of the p values obtained in t test comparison. Protein classes are highlighted. Note the strong presence of mitochondrial proteins in 3D samples.
(B) Scatterplots demonstrating the mean log10 normalized spectral counts of 2D and 3D hiPSC-CMs. Proteins enriched in 3D are located above the line of best fit and vice versa for 2D. Protein classes are highlighted.
(C) Volcano plot showing main cardiac proteins highlighted in green.
(D) Examples of mitochondrial protein spectral count analysis involved in fatty acid oxidation (FAO) and respiration (data extracted from A). Samples (n = 3 per group) prepared from one experiment.
∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001, EHT versus 2D (two-way ANOVA plus Bonferroni post test); error bars show means ± SEM. Related data are depicted in Figure S2.