Figure 3.
Comparison of the Mitochondrial Proteome between 2D, 3D, and Non-failing Human Hearts
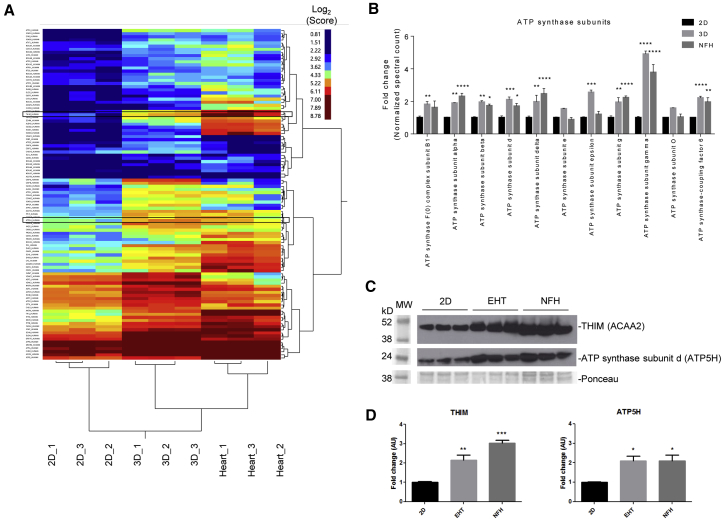
(A) Clustering analysis of mitochondrial proteins for 2D and 3D hiPSCs and non-failing heart samples. Expression levels are depicted as a color code ranging from blue (low expression) to red (high expression).
(B–D) Further analysis was performed for a selection of proteins involved in fatty acid oxidation and respiratory chain. (B) Proteome data of ATP synthase subunits. (C) Western blot analysis of 2D, 3D, and NFH for mitochondrial proteins such as 3-ketoacyl-CoA thiolase (ACAA2) and ATP synthase subunit d (ATP5H) (framed in A) in comparison with Ponceau staining. (D) Quantification of western blot analysis in (C).
Samples (n = 3 per group) prepared from one experiment. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.001 versus 2D (one-way ANOVA plus Bonferroni post test); error bars show means ± SEM. Related data are depicted in Figure S3.