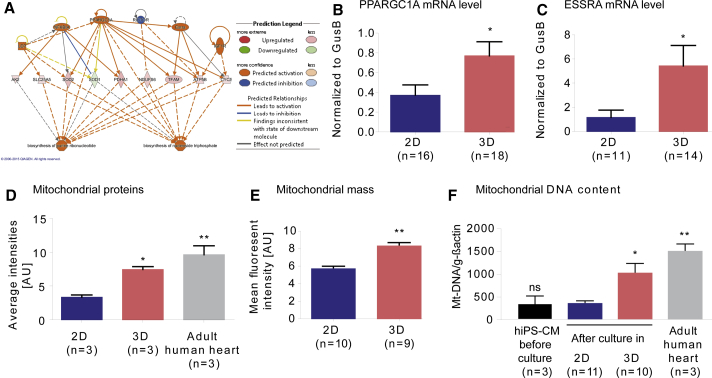
Figure 4.
Expression of Mitochondrial Biogenesis-Related PGC-1α Signaling Pathway and Mitochondrial Protein, Mass, and DNA Content in hiPSC-CMs
(A) Analysis of “Regulator Effects” by IPA of proteins significantly different between 2D and 3D: highest consistency score for effector network involved in the biosynthesis of purine ribonucleotides and nucleoside triphosphates and directed by PPARGC1/PGC-1α and ESSRA, among other proteins.
(B and C) qRT-PCR for PPARGC1/PGC-1α (B) and ESSRA (C).
(D) Average intensity (normalized spectral counts as a measure for relative protein abundance) between samples of mitochondrial proteins detected by quantitative proteomics in hiPSC-CMs cultured in 2D or 3D compared with non-failing heart; protein must be identified in at least 2 out of 9 samples.
(E) Quantification of flow cytometry with MitoTracker Green FM of 2D versus 3D CMs.
(F) PCR amplification of genomic DNA for the mitochondrial encoded NADH dehydrogenase (Mt-ND1/2), normalized to the nuclear encoded gene actin, human heart samples (1× non-failing, 2× terminal heart failure).
n denotes biological replicates, ≥3 independent experiments of both control cell lines. ∗p < 0.05 and ∗∗p < 0.01 versus 2D (two-tailed unpaired t test, B, C, and E; error bars show means ± SEM). ∗p < 0.05 and ∗∗p < 0.01 versus 2D (one-way ANOVA plus Bonferroni post test, D and F; error bars show means ±SEM; ns, not significant). Related data are depicted in Figure S4.