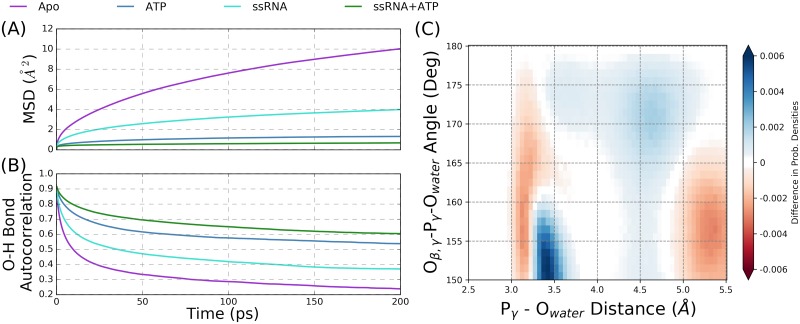
Fig 4. Water dynamics and positioning within the NTPase active site.
(A) Mean square displacement (MSD) and (B) O-H bond autocorrelation metrics for the Apo, ATP, ssRNA, and ssRNA+ATP simulations that describe the translational and rotational motions of waters within the active site. (C) The difference between the ssRNA+ATP and ATP probability densities of water positions within the NTPase active site, projected onto the Oβ,γ-Pγ-Owat angle and Pγ-Owat distance. These axes are used to project water positions into catalytically relevant space relative to the ideal position of a lytic water in the hydrolysis reaction.