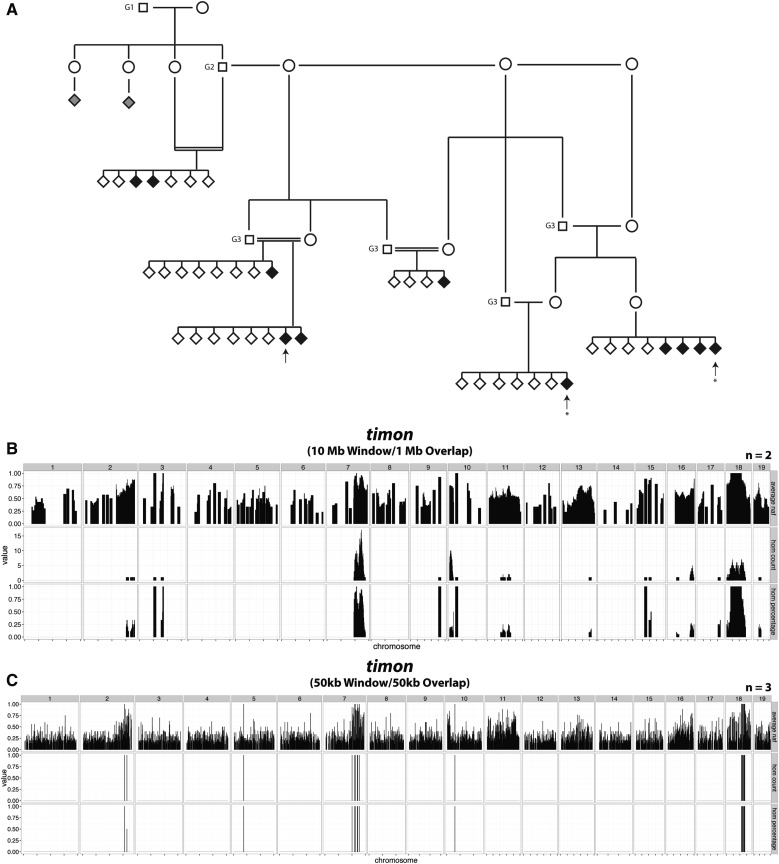
Figure 4.
Pedigree and homozygosity mapping of timon indicates autosomal recessive inheritance and a causal locus on mouse chromosome 18. (A) The timon pedigree indicates two distinct branches: one that is more closely related to the G1 and one that is farther removed. Two individuals (gray diamonds) exhibited more severe craniofacial and limb phenotypes, and were excluded as affected by genotyping. Therefore, these two embryos represent a mutant phenotype distinct from timon. Arrows, genomic DNA samples selected for WGS. Asterisks: two individuals used to map the critical interval in (B). (B) The timon causal locus was mapped to chromosome 18 using WGS data from two of the three samples submitted. A peak on chromosome 7 was eliminated by genotyping affected individuals across the critical interval for candidate ENU-induced variants (Table S4 in File S1). (C) Reducing the window and overlap size to 50 kb in all three individuals’ mapping analysis revealed that the third sample also possessed the homozygous peak on chromosome 18. ENU, N-ethyl-N-nitrosourea; WGS, whole-genome sequencing.