Figure 4. Tuning is both promoter- and allele-specific.
(A) Cell counts for synprom-URA3-mRuby/PADH1-DHFR-GFP cells in liquid ura-/6AU5 media. Colors correspond to different biological replicates started on different days. Arrows indicate two timepoints from each strain for which fluorescence cumulative distribution functions (CDFs) are shown below. Error bars for cell counts show central 95% credible intervals. (B) Flow cytometry cumulative distributions of fluorescence levels for URA3-mRuby and DHFR-GFP during uracil starvation. In each CDF a given timepoint (solid line) is compared to the distribution present for cells in logarithmic growth in SC+glu (rich) media (dashed lines). The values shown are log2 ratios to the median value of cells growing exponentially in SC+glu. GFP signals are shown in green and mRuby signals in red. (C) Analogous to A, but we consider cells where synprom drives both URA3-mRuby and DHFR-GFP. (D) Analogous to B, but for cells with synprom driving both URA3-mRuby and DHFR-GFP.
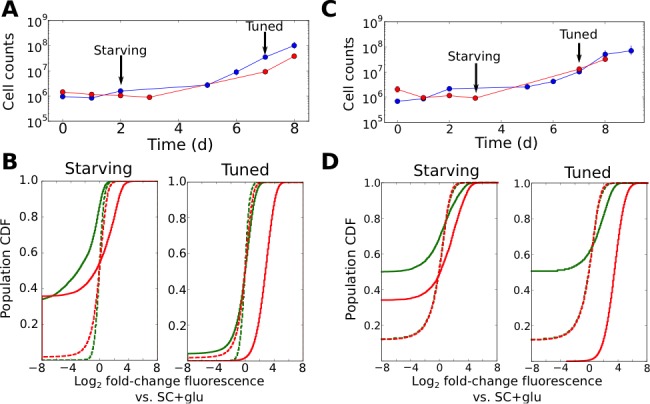
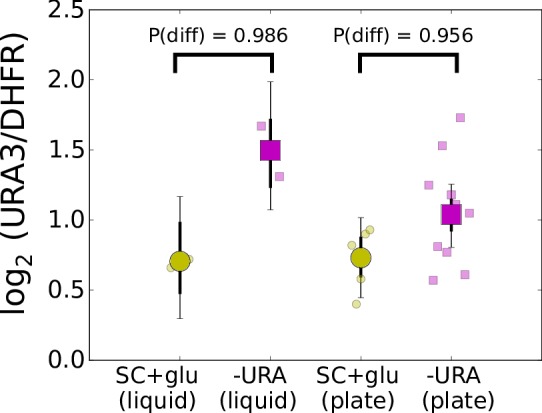
Figure 4—figure supplement 1. Promoter-specific stochastic tuning of URA3 expression by native promoters in S. cerevisiae.
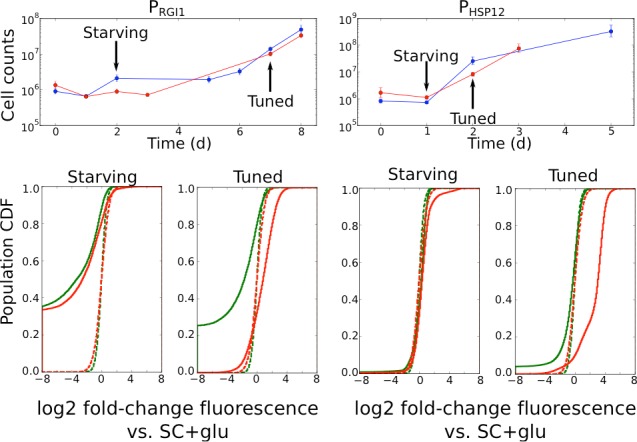
Figure 4—figure supplement 2. Local tuning of URA3 expression.