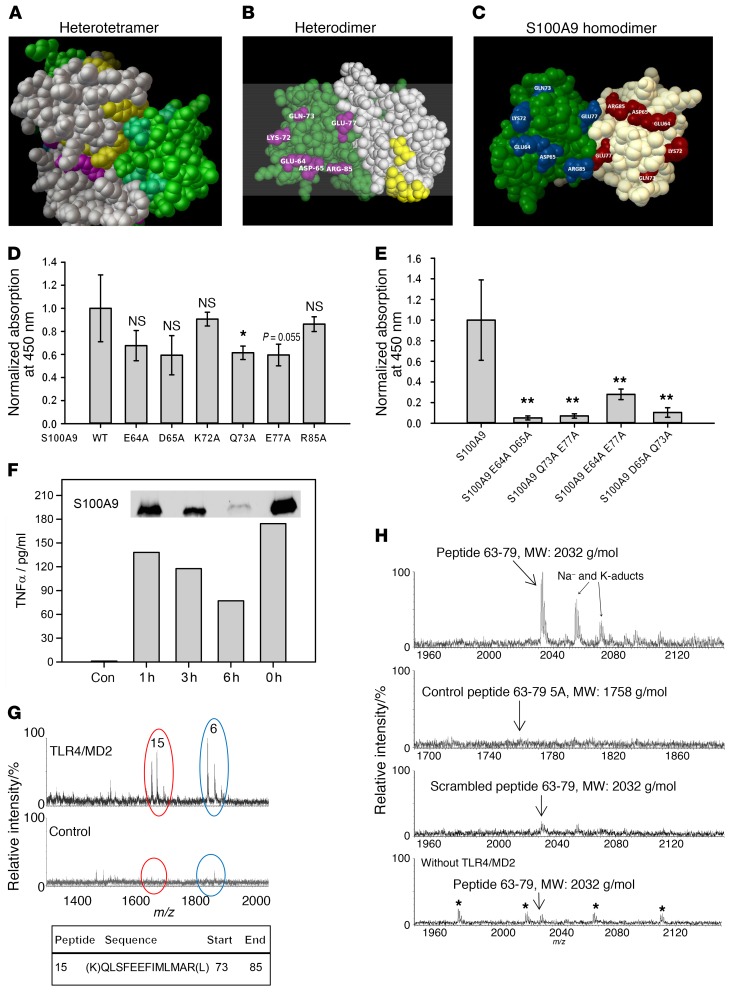
Figure 5. Identification of the S100A9-binding site on TLR4/MD2.
(A–C) The PDB files of the S100A8/S100A9 tetramer (1XK4, A) and S100A9 dimer (1IRJ, C) were retrieved from the RSCB PDB website. (B) 1XK4 was modified containing only the E (S100A8, gray) and G (S100A9, green) chains resembling 1 heterodimer. Masked aa in the tetramer were analyzed by Cluspro. (B, yellow- or cyan-labeled aa for S100A8 or S100A9). Labeled S100A9 aa were chosen for mutation studies (C). (D) Single mutated S100A9 was analyzed for TLR4/MD2 binding compared with WT S100A9. All S100A9 mutants showed reduced binding to TLR4/MD2. (E) Double-mutated S100A9 was analyzed for binding to TLR4/MD2 as shown in D and showed significantly reduced binding properties compared with WT S100A9. Data represent mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01, 1-way ANOVA. (F) Monocytes were stimulated for 4 hours with intact S100A9 (0 h = no trypsinization) or the fragment mixture, and TNF-α levels were quantified by ELISA. Western blot shows remaining intact S100A9 (insert). One representative of 3 independent experiments is shown. (G) Tryptic fragments of hS100A9 (F) were analyzed for TLR4/MD2 binding, and peptide 15 corresponding to aa 73–85 showed specific interaction with TLR4/MD2, as detected by nanoUPLC/ESI-Q-TOF MS/MS (m/z 1614). Peptide 6 (NIETIINTFHQYSVK) was also detected in the control setting without TLR4/MD2 and reflects unspecific binding. (H) Peptides comprising aa 63–79 of S100A9 (MEDLDTNADKQLSFEEF, MW: 2032 g/mol), control peptides 63–79 5A (MAALDTNADAALSFAEF, MW: 1758 g/mol) or a scrambled peptide (DSLEMTEENLADQFKDF, MW: 2032 g/mol) were investigated for TLR4/MD2 binding as shown in G. Unspecific binding was analyzed by using peptides 63–79 without TLR4/MD2. Only S100A9 peptides 63–79 showed binding to TLR4/MD2 (3 independent experiments). Asterisks indicate polyethylene glycol impurities.